Exosomal miR-92b-5p regulates N4BP1 to enhance PTEN mono-ubiquitination in doxorubicin-resistant AML
Abstract
Aim: Doxorubicin, pivotal for acute myeloid leukemia (AML) treatment, often succumbs to resistance, impeding therapeutic success. Although exosomal transfer is linked to chemoresistance, the detailed role of exosomal miRNAs in doxorubicin resistance remains incompletely understood.
Methods: We employed miRNA sequencing to delineate the profile of exosomal miRNAs in doxorubicin-resistant K562/DOX cells and AML patients. Subsequently, qPCR was utilized to scrutinize the expression of exosomal miR-92b-5p in these resistant cells and AML patients. A dual-luciferase reporter assay was conducted to elucidate the direct binding of miR-92b-5p to NEDD4 binding protein 1 (N4BP1). Furthermore, interactions between N4BP1 and NEDD4, as well as between NEDD4 and PTEN, were investigated by co-immunoprecipitation (Co-IP). Meanwhile, the ubiquitination of PTEN was also examined by Co-IP. Western blot analysis was applied to assess the expression levels of N4BP1, NEDD4, PTEN, RAD51, and proteins associated with the PI3K-AKT-mTOR pathway. Gain- and loss-of-function studies were conducted to ascertain the functional role of miR-92b-5p in doxorubicin resistance by using miR-92b-5p-mimic and miR-92b-5p-inhibitor transfections.
Results: Our study found exosomal miR-92b-5p was upregulated both in doxorubicin-resistant cells and AML patients. Moreover, miR-92b-5p targets N4BP1, promoting NEDD4-mediated mono-ubiquitination of PTEN. This alters PTEN’s subcellular localization, promoting nuclear PTEN and reducing cytoplasmic PTEN, which in turn leads to increased RAD51 for DNA repair and activation of the PI3K-AKT-mTOR pathway for cell proliferation, contributing to doxorubicin resistance.
Conclusion: Our study reveals a novel mechanism of doxorubicin resistance mediated by exosomal miR-92b-5p and provides potential therapeutic targets for overcoming drug resistance in AML.
Keywords
INTRODUCTION
Acute myeloid leukemia (AML) is a hematological malignancy marked by the proliferation of abnormal white blood cells, severely impacting blood production. Doxorubicin, a widely utilized anthracycline antibiotic, is a cornerstone of AML chemotherapy due to its ability to intercalate DNA and generate free radicals, thereby inhibiting DNA replication and transcription. Despite advances in chemotherapeutic regimens, the emergence of doxorubicin resistance remains a significant challenge, undermining treatment efficacy and leading to poor patient outcomes[1]. There is a critical need to understand the molecular underpinnings of this resistance to develop novel therapeutic strategies.
MicroRNAs (miRNAs), which are 18-24 nucleotide non-coding RNAs, interact with the miRNA-induced silencing complex (miRISC) to target and regulate the expression of genes involved in the cell cycle, proliferation, apoptosis, DNA damage repair, and drug metabolism, thereby influencing drug resistance in leukemia[2-4]. For example, miR-128 overexpression led to increased DNA damage, which subsequently reduced cell viability and enhanced the cells’ sensitivity to etoposide and doxorubicin[5]. Additionally, miR-133 has been reported to boost doxorubicin sensitivity in Evi1-overexpressing cells by promoting apoptosis but has no effect in Evi1-deficient cells[6]. Furthermore, miR-130a knockdown increased the sensitivity of AML cells to doxorubicin by decreasing cell viability[7]. Nevertheless, there remains a subset of AML patients with doxorubicin resistance that cannot be attributed to the known miRNAs and their mechanisms. Therefore, it is imperative to discover novel miRNAs and explore their underlying mechanisms, which could provide fresh insights for the diagnosis, treatment, and prognostic evaluation of AML.
Exosomes are tiny vesicles released by cells and packed with miRNAs and other small molecular cargo[8]. Exosomes facilitate the transfer of miRNAs between cells, thereby fostering cancer progression and drug resistance by altering drug targets, reducing intracellular drug concentrations, and regulating cell cycle, apoptosis, and DNA damage repair in recipient cells[9-14]. For example, exosomal miR-365 from imatinib-resistant chronic myeloid leukemia (CML) K562/G01 cells confer imatinib resistance to sensitive K562 cells by inhibiting Bax and caspase-3[15]. It has been reported that exosomal miR-19b and miR-20a in multidrug-resistant AML HL-60/ADR cells spread multidrug resistance among sensitive HL-60 cells upon co-culture[16]. Despite these findings, there is a notable gap in research specifically investigating the role of exosomal miRNAs in doxorubicin resistance in AML patients and cell lines. Therefore, this study mainly investigated the exosomal miRNA in doxorubicin-resistant AML, explored and verified its target genes and pathways, thereby providing new targets and ideas for the treatment of doxorubicin-resistant AML.
METHODS
Study participants
Blood samples were collected from AML patients before the initiation of chemotherapy. The patient cohort comprised 5 patients with newly diagnosed AML who responded to chemotherapy and 5 patients with refractory AML who did not respond to the doxorubicin-included regimen between January 2022 and December 2023 [Supplementary Table 1]. The diagnosis and relapse/refractory status of AML were proved by morphology examination, flow cytometry analysis, karyotype analysis, and molecular detection.
Cell culture and transfection
Human leukemia cell K562 and its doxorubicin-resistant cell line K562/DOX were obtained from the Cell Center of Xiangya Medical College and were authenticated by STR profiling (Genetic Testing Biotechnology, Suzhou, China). K562 cells were cultured in RPMI-1640 culture medium (Gibco, USA) with 10% FBS (11012-8611, EVERYGREEN, Zhejiang, China), K562/DOX cells were cultured with 1 μM doxorubicin (KGA8184, KeyGen, Jiangsu, China) additionally for maintaining resistance and were cultured in doxorubicin-free culture medium before experiment.
After cell confluence reached 70%-80%, K562 or K562/DOX cells (1 × 106 cells/well) were inoculated into 6-well plates and transfected with 100 nM miR-92b-5p mimic or inhibitor (R10034.9, RIBO, Guangzhou, China) for 6 h and cultured for 48 h.
Exosome isolation and identification
After cell confluence reached 70%-80%, K562 or K562/DOX cells were washed twice with PBS (G4202, Servicebio, Wuhan, China) and cultured in RPMI-1640 culture medium with 5% exosome-depleted FBS (Cat#EXO-FBS-50A-1, SBI, Shanghai, China) for 24 h. The cell suspensions were centrifuged at 300 g for
The patient’s blood samples were collected with an EDTA anticoagulant tube and centrifuged at 1,500 g for 20 min at 4 °C. The supernatants were collected and centrifuged at 3,000 g for 15 min at 4 °C. Exosomes were isolated using Exosome Purification Kit-Exosupur (Cat#Echo9101A-10ml, Echo Biotech, Beijing, China) following the manufacturer’s protocols.
Exosome nanoparticle size distribution was confirmed by Nanoparticle Tracking Analysis [Supplementary Figure 1A]. The morphology of the exosome was identified by transmission electron microscopy (TEM) at the Institute of Pathology, Xiangya Medical College of Central South University [Supplementary Figure 1B]. The exosome markers calnexin, Hsp90, CD63, and Alix were detected by Western blots [Supplementary Figure 1C].
Small RNA extraction and sequencing
Small RNA was extracted using the miRNeasy Mini Kit (Cat. No. 217004, Qiagen, Germany) following the manufacturer’s protocols. Subsequently, small RNA libraries were constructed and sequenced on Illumina HiSeq2500 platforms by Echo Biotech company (Beijing, China). MiRNA libraries were constructed using QsRNA-seq[17], and unique molecular identifiers (UMIs) were employed to avoid bias during PCR. The miRNA expression levels were normalized using the TPM algorithm.
Doxorubicin resistance-related miRNAs and target gene functional annotation
Differential expression analysis of two groups was performed using the edgeR R package (3.12.1)[18].
MiRanda[19] and RNAhybrid[20] were used for miRNAs target gene prediction, and BLAST software was used to compare the predicted target gene sequence with databases such as NR[21], Swiss Prot[22], gene ontology (GO)[23], COG[24], Kyoto Encyclopedia of Genes and Genomes (KEGG)[25], KOG[26], and Pfam[27] for the miRNA target gene functional annotation. The topGO R package was utilized to perform GO enrichment analysis on the target genes of differentially expressed miRNAs. KOBAS[28] was employed to assess the statistical enrichment of differentially expressed miRNA target genes in KEGG pathways.
q-PCR
The Pubmed GENE database (https://pubmed.ncbi.nlm.nih.gov) and miRBase database
Protein extraction and western blots
The cells were treated with the RIPA buffer (KGP702, KeyGen, Jiangsu, China) to obtain total protein, and the nuclear and cytoplasmic proteins were extracted using a cytoplasmic and nuclear protein extraction kit (KGP150, KeyGen, Jiangsu, China). The protein was loaded onto a 10% SDS-PAGE gel and subsequently transferred to a PVDF membrane. The membrane was blocked with 5% BSA in TBST and incubated with primary antibodies overnight at 4 °C. Afterward, HRP-linked anti-rabbit IgG secondary antibody (BL003A, Biosharp, Beijing, China) was applied at a 1:10,000 dilution and incubated at 25 °C for 1 h. The protein bands were visualized using ECL reagents (BL520A, biosharp, Beijing, China). The Tublin, PCNA, and GAPDH protein levels were used as internal controls. Primary antibodies are listed in Supplementary Table 3.
Dual-luciferase assay
We purchased psiCHECK2 vectors with N4BP1 3’-UTRs wild-type (WT) or mutant (MUT) sequences from Changsha Abiowell Biotechnology (Changsha, China). 293T cells were transfected with one of the above vectors along with the miR-92b-5p mimic or mimic-NC using Lipofectamine 2000 (Thermo Fisher Scientific, USA) for 72 h. Luciferase activities were measured using the TransDetect Double-Luciferase Reporter Assay Kit (Cat#: FR201, TransGen, China).
Co-immunoprecipitation experiment
The co-immunoprecipitation (Co-IP) experiment was conducted using Pierce Classic magnetic immunoprecipitation kit (Cat#:88804, Thermo Fisher Scientific, USA). Proteins were immunoprecipitated from cell lysates with primary antibodies (anti-NEDD4 antibody, anti-UB antibody, anti-PTEN antibody, and anti-IgG antibody are listed in Supplementary Table 3). Protein complexes were bound to Protein A/G Magnetic Beads at 25 °C for 1 h, the beads were washed twice with wash buffer and once with ddH2O, and the protein complexes were eluted with low-pH buffer and analyzed by western blots. The secondary antibody was replaced by HRP-conjugated anti-rabbit IgG, Light Chain Specific (SA00001-7L, Proteintech, Wuhan, China); the remaining steps are as described above.
Immunofluorescence
K562 or K562/DOX cells (2 × 105 cells/well) were inoculated into 24-well plates and transfected with 100 nM miR-92b-5p mimic or inhibitor for 6 h, followed by a 48-hour culture period. The cells were washed twice with PBS and resuspended in 0.1 mL PBS (2-5 × 106 cells/mL). Shi-fixTM cover-slips (#SB-Shifix25, Shikhar Biotech, Lalitpur, Nepal) were placed in a 12-well plate. The cell suspension was added directly onto the Shi-fixTM cover-slips. The samples were incubated at room temperature for 30 min, after which unbound cells were gently removed by washing with 1 mL PBS. The Shi-fixTM cover-slips were fixed, permeabilized, and then incubated with primary antibodies (bs-0748R, Bioss Biotechnology, listed in Supplementary Table 3) overnight at 4 °C and HRP-linked anti-rabbit IgG secondary antibody (AFIHC003, Aifang biological, Changsha, China) for 30 min at 25 °C in 12-well plate. Afterward, the Shi-fixTM cover-slips were incubated with TYR-570 reagents (AFIHC027, Aifang biological, Changsha, China) and DAPI (AFIHC044, Aifang biological, Changsha, China) and visualized in Digital Pathology Scanner (KF-FL-020, KFBIO).
CCK-8 assay
A total of 1 × 104 cells were cultured with different concentrations of doxorubicin and for 48 h in 96-well plates. The CCK-8 solution (A311, Vazyme, Nanjing, China) was added; after incubation for 4 h, the OD value was measured at 450 nm wavelength using the multimode plate reader (EnVision Xcite, USA).
Apoptosis detection by flow cytometry
A total of 5 × 105 cells were cultured with 5 μM doxorubicin and for 4 h. After treatment, cells were collected and gently washed with cold PBS twice. Then, cells were resuspended in 500 μL 1× binding buffer, 5 μL Annexin V-FITC, and 5 μL PI using Annexin V-FITC/PI Apoptosis Detection Kit (KGA1102-50, KeyGen, Jiangsu, China). The stained cells were incubated in the dark for 5 min at room temperature and then analyzed using a flow cytometer (DxP AthenaTM, Cytek Biosciences).
Statistical methods
Triplicates of all experiments were performed at least. For the data in this study, all were presented in the form of mean ± standard deviation. Data comparison between two groups was performed using a t-test, while comparisons among multiple groups were conducted using one-way ANOVA. Statistical significance was set at P < 0.05. (*P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001, n = 3).
RESULTS
Doxorubicin resistance-related exosomal miRNA in cell lines and AML patients
To explore the mechanisms underlying doxorubicin resistance transferred by exosomes, we investigated exosomal miRNA profiles in doxorubicin-resistant cells and chemoresistant AML patients through high-throughput miRNA sequencing. The results revealed 77 exosomal miRNAs were upregulated and 41 exosomal miRNAs were downregulated in the doxorubicin-resistant K562/DOX cells compared with doxorubicin-sensitive K562 cells [Figure 1A and B], 64 exosomal miRNAs were upregulated and 36 exosomal miRNAs were downregulated in chemoresistant AML patient [Supplementary Figure 2A and B]. The GO analysis suggested that the predicted target genes of differentially expressed exosomal miRNAs in doxorubicin-resistant K562/DOX cells may be associated with biological processes such as “cell aggregation”, cellular components like “extracellular matrix part collagen”, and molecular functions including “protein binding transcription factor activity”, “translation regulator activity”, and “chemorepellent activity” [Figure 1C]. To further elucidate the biological roles of these predicted target genes in doxorubicin-resistant K562/DOX cells, a KEGG analysis was conducted. This analysis identified key pathways potentially involving these target genes, with “Protein processing in endoplasmic reticulum”, “Olfactory transduction”, “Lysosome”, “Endocytosis”, and “Transcriptional misregulation in cancer” emerging as the top five [Figure 1D]. Furthermore, KEGG enrichment analysis indicated significant associations between the predicted target genes and pathways such as “Basal cell carcinoma”, “AML”, “Amoebiasis”, “Glycosaminoglycan biosynthesis - heparan sulfate/heparin”, and “Choline metabolism in cancer” [Figure 1E]. Notably, the “AML” pathway showed the highest enrichment factor, suggesting a considerable level of involvement. This result implies a potential link between the differential expression of exosomal miRNAs in K562/DOX cells and the development of AML.
Figure 1. Comprehensive analysis of differentially expressed exosomal miRNAs between doxorubicin-resistant K562/DOX cells and the doxorubicin-sensitive K562 cells and their target genes. (A) Volcano plot of differentially expressed exosomal miRNAs; (B) Cluster map of differentially expressed exosomal miRNAs; (C) GO analysis of differentially expressed exosomal miRNAs target genes; (D) KEGG analysis of differentially expressed exosomal miRNAs target genes; (E) KEGG pathway enrichment of differentially expressed exosomal miRNAs target genes. miRNAs: MicroRNAs; GO: gene ontology; KEGG: Kyoto Encyclopedia of Genes and Genomes.
Exosomal miR-92b-5p is related to doxorubicin resistance in AML patients and cell lines
Based on exosomal miRNA-seq, we identified 10 miRNAs that exhibited increased expression levels in both doxorubicin-resistant K562/DOX cells and in chemoresistant AML patients, as detailed in Supplementary Table 4. We focused on the top five - miR-92b-5p, miR-671-5p, miR-378a-3p, miR-378c, and miR-378i - for validation. Firstly, we conducted quantitative PCR to measure the expression levels of above 5 miRNAs in exosomes derived from 5 chemotherapy-resistant AML patients and 5 chemotherapy-sensitive AML patients, as well as from K562/DOX and K562 cells. The results confirmed the significantly higher exosomal miR-92b-5p levels in chemoresistant AML patients (P < 0.0106, Figure 2A) and in K562/DOX cells (P < 0.017, Figure 2B). Exosomal miR-378a-3p and miR-378c also showed increased expression in chemoresistant AML patients and K562/DOX cells [Supplementary Figure 3A-D], while exosomal miR-671-5p and miR-378i elevated in K562/DOX cells but not in AML resistant patients [Supplementary Figure 3E-H]. The validation studies have identified exosomal miR-92b-5p as the most highly expressed miRNA in K562/DOX cells, with the most marked differences; therefore, we have selected exosomal miR-92b-5p as a potential biomarker for further investigation. Then, we discovered a significant upregulation of miR-92b-5p in K562/DOX cells compared to K562 cells. (P < 0.0001, Figure 2C). To verify the correlation between miR-92b-5p and doxorubicin resistance, we conducted transfections with miR-92b-5p mimics or inhibitors to regulate its expression within cells. Overexpression via miR-92b-5p mimics resulted in enhanced doxorubicin resistance in K562 cells (K562-mimic), evidenced by an increase in the IC50 for doxorubicin, whereas the inhibition of miR-92b-5p in K562/DOX cells (K562/DOX-inhibitor) corresponded with a decrease in doxorubicin resistance, as reflected by a reduced IC50 [Figure 2D]. Moreover, we found that ABCB1 and ABCG mRNA levels are elevated in K562/DOX cells, but miR-92b-5p does not affect their expression in K562 or K562/DOX cells [Supplementary Figure 4].
Figure 2. Exosomal miR-92b-5p is related to doxorubicin resistance in AML patients and cell lines. (A) The expression of exosomal miR-92b-5p in AML patients; (B) The expression of exosomal miR-92b-5p in K562 and K562/DOX cells; (C) The expression of miR-92b-5p in K562 and K562/DOX cells; (D) The IC50 value of doxorubicin in K562 and K562/DOX cells. *P < 0.05, **P < 0.01, ****P < 0.0001, n = 3. AML: Acute myeloid leukemia.
miR-92b-5p binds to and suppresses N4BP1 expression in doxorubicin-resistant leukemia cells
N4BP1 was identified as a target gene for miR-92b-5p following bioinformatics analysis with TargetScan
Figure 3. miR-92b-5p binds to and suppresses N4BP1 expression in doxorubicin-resistant leukemia cells. (A) Binding site of miR-92b-5p to N4BP1 mRNA 3’-UTR; (B) The relative Luciferase activity in 293T cells co-transfected with miR-92b-5p mimics or miR-NC and N4BP1-3’-UTR-WT or N4BP1-3’-UTR-MUT; (C) The expression of N4BP1 mRNA in K562 and K562/DOX cells; (D) The expression of N4BP1 mRNA in K562 and K562/DOX cells transfected with miR-92b-5p mimics or inhibitors; (E-G) The expression of N4BP1 protein in K562 and K562 /DOX cells transfected with miR-92b-5p mimics or inhibitors. *P < 0.05, **P < 0.01, ***P < 0.001, n = 3. N4BP1: NEDD4 binding protein 1.
NEDD4-mediated PTEN ubiquitination and its association with N4BP1 in doxorubicin-resistant leukemia cells
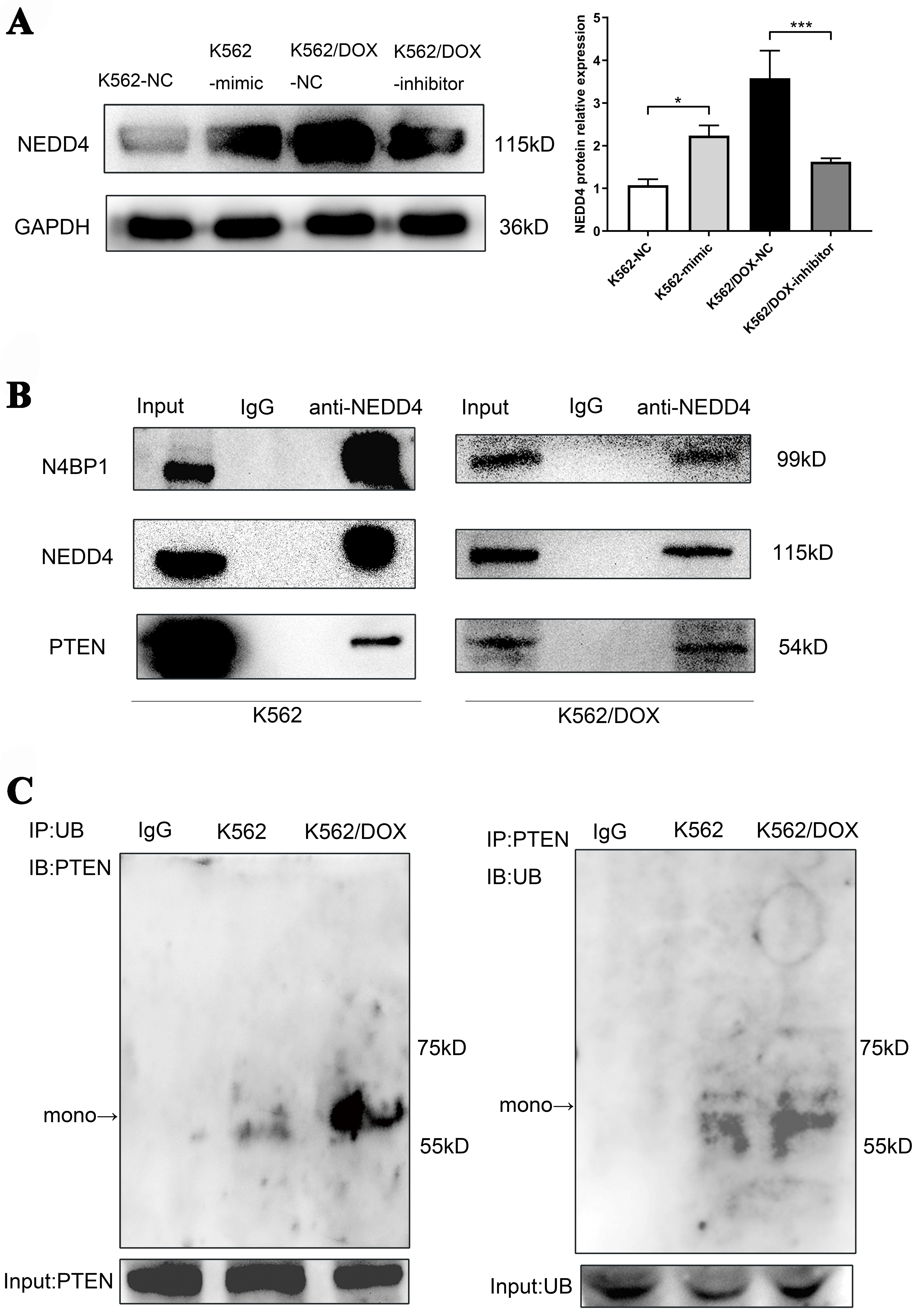
To confirm our hypothesis that N4BP1 participates in PTEN ubiquitination mediated by NEDD4, potentially affecting the doxorubicin resistance in leukemia cells, we assessed the expression of NEDD4 and the levels of PTEN ubiquitination in K562 and K562/DOX cells. Our findings indicate that overexpression of miR-92b-5p in K562 cells increased NEDD4 protein levels, whereas its inhibition in K562/DOX cells led to a decrease in NEDD4 expression [Figure 4A], which is consistent with the higher level of PTEN mono-ubiquitination assessed by Co-IP in K562/DOX cells as we expected [Figure 4B]. Furthermore, we performed Co-IP experiments using NEDD4 antibodies, and confirmed that NEDD4 is binding to N4BP1 and PTEN in both K562 and K562/DOX cells [Figure 4C].
Figure 4. Analysis of NEDD4 expression, PTEN ubiquitination, and protein interactions in K562 and K562/DOX cells. (A) The expression of NEDD4 protein in K562 and K562/DOX cells transfected with miR-92b-5p mimics or inhibitors; (B) The expression of mono-ubiquitinated PTEN protein in K562 and K562/DOX cells; (C) NEDD4 and N4BP1, NEDD4 and PTEN endogenous Co-IP in K562 cells and K562/DOX cells. *P < 0.05, ***P < 0.001, n = 3. N4BP1: NEDD4 binding protein 1; Co-IP: co-immunoprecipitation.
miR-92b-5p regulates RAD51 expression and PI3K-AKT-mTOR pathway via modulating PTEN subcellular localization
Given the influence of PTEN mono-ubiquitination on its nuclear distribution[29], we subsequently explored the influence of miR-92b-5p on the subcellular expression levels of PTEN. Western blots results revealed that compared to the transfection of miRNA-mimic negative control (NC), transfection with miR-92b-5p-mimic in K562 cells increased nuclear PTEN protein, alongside decreased total and cytoplasmic PTEN protein. Consistently, compared to miR-inhibitor NC, miR-92b-5p inhibitor triggered a reduction in nuclear PTEN protein in K562/DOX cells, coupled with upregulated total and cytoplasmic PTEN protein
Figure 5. miR-92b-5p regulates RAD51 expression and PI3K-AKT-mTOR pathway via modulating PTEN subcellular localization. (A and B) The expression and relative expression of PTEN in total protein, cytoplasmic and nuclear proteins in K562 and K562/DOX cells transfected with miR-92b-5p mimics or inhibitors; (C) Immunofluorescence of PTEN in K562 and K562/DOX cells transfected with miR-92b-5p mimics or inhibitors; (D and E) The expression and relative expression of RAD51 in total protein and nuclear proteins in K562 and K562/DOX cells transfected with miR-92b-5p mimics or inhibitors; (F-H) The expression and relative expression of PI3K-AKT-mTOR pathway in total protein and cytoplasmic protein in K562 and K562/DOX cells transfected with miR-92b-5p mimics or inhibitors. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001, n = 3.
miR-92b-5p affects doxorubicin resistance through cell apoptosis and proliferation
Due to the activation of the PI3K-AKT-mTOR pathway promoting cell proliferation, and the enhancement of DNA damage repair by nuclear RAD51, thereby reducing cell apoptosis, we detected the effect of miR-92b-5p on the proliferation and apoptosis levels of K562 and K562/DOX cells. The CCK-8 assays demonstrated that the proliferation of K562 cells overexpressing miR-92b-5p (K562-mimic) was significantly higher than that of the control cells [Figure 6A]. Conversely, the proliferation of K562/DOX cells with miR-92b-5p suppression (K562/DOX-inhibitor) was notably reduced compared to the control cells [Figure 6B]. Western blot analysis revealed that the apoptosis proteins casepase-3 and cleaved-PARP1 in K562-mimic cells were reduced compared to the control cells [Figure 6C and D], whereas these proteins were elevated in K562/DOX-inhibitor cells [Figure 6C and E]. Flow cytometry corroborated these findings, showing that the apoptosis rate of K562-mimic cells was decreased compared to the control [Figure 6F and G], while the rate in K562/DOX-inhibitor cells was increased [Figure 6F and H]. Collectively, these findings substantiate the role of miR-92b-5p in augmenting cell proliferation and attenuating cell apoptosis.
Figure 6. miR-92b-5p promotes cell proliferation and attenuates cell apoptosis. (A) The cell viability of K562 cells transfected with miR-92b-5p mimics; (B) The cell viability of K562/DOX cells transfected with miR-92b-5p inhibitors; (C-E) The expression and relative expression of capase-3 and cleaved-PARP1 proteins in K562 and K562/DOX cells transfected with miR-92b-5p mimics or inhibitors; (F-H). Apoptosis rate in K562 and K562/DOX cells transfected with miR-92b-5p mimics or inhibitors. *P < 0.05, **P < 0.01, n = 3.
DISCUSSION
In this study, we conducted a comparative analysis of exosomal miRNAs between the parental K562 cells and the doxorubicin-resistant K562/DOX cells. MiRNAs-Seq analysis revealed a significant upregulation of 77 exosomal miRNAs in the K562/DOX cells, with miR-92b-5p exhibiting the most pronounced increase. Through GO and KEGG analyses, we identified that these miRNAs target genes involved in critical biological processes such as “biological adhesion”, “growth”, “cell aggregation”, “Transcriptional misregulation in cancer”, and “AML”. Further studies confirmed that miR-92b-5p directly targets N4BP1 mRNA, reducing its mRNA and protein levels. This downregulation of N4BP1 increases NEDD4 expression, promoting PTEN mono-ubiquitination and nuclear accumulation, while decreasing its total and cytoplasmic levels. Elevated nuclear PTEN induces RAD51 upregulation, enhancing DNA repair, while reduced cytoplasmic PTEN activates the PI3K-AKT-mTOR pathway, driving cell proliferation and inhibiting apoptosis. We propose that RAD51 and the PI3K-AKT-mTOR pathway are key downstream targets of miR-92b-5p in mediating doxorubicin resistance in leukemia.
The GO analysis revealed that the differential miRNA target genes in doxorubicin-resistant K562/DOX cells were enriched in “biological adhesion”, “growth”, “cell aggregation”, and were decreased in “cell killing”. An increase in “biological adhesion” and “cell aggregation” suggested the intercellular interactions and aggregation behaviors may increase in doxorubicin-resistant leukemia cells; this finding is consistent with that “biological adhesion” might correlate with doxorubicin resistance in leukemia[31], and “cell aggregation” is thought to involved in drug resistance of AML cells [32]. An increase in “growth” suggested that cell growth and proliferation might play a crucial role in doxorubicin-resistant leukemia. Moreover, a decrease in “cell killing” indicated that cell death and apoptosis in doxorubicin-resistant leukemia are diminished, which decreases the sensitivity of leukemia cells to cytotoxic stimuli, thereby promoting leukemia cell survival and inducing resistance to chemotherapy. In addition, KEGG analyses indicated these differential miRNAs target genes are enriched in “Transcriptional misregulation in cancer”, “MicroRNAs in cancer”, “Proteoglycans in cancer”, and “Pathways in cancer” in terms of “Human Diseases”. Moreover, KEGG pathway enrichment analyses identified “AML” as the most significant pathway, with the highest enrichment factor, which implied that these miRNAs could be involved in the development of cancer, especially in AML. It is worth mentioning that KEGG analyses indicated differential miRNAs target genes might also be involved in “Dilated cardimyopathy”, which suggests that these miRNAs may regulate cardiac muscle function and participate in the process of cardiac remodeling and functional deterioration. This result is consistent with that exosomal miR-92b-5p may serve as potential biomarkers for acute heart failure caused by dilated cardiomyopathy[33].
Based on exosomal miRNA-seq, we identified 10 miRNAs that exhibited increased expression levels in both doxorubicin-resistant K562/DOX cell lines and in chemoresistant AML patients. Quantitative PCR validated that miR-92b-5p, miR-378a-3p, and miR-378c exhibited increased expression in K562/DOX cells and in resistant AML patients. Consistent with our findings, research revealed that chemotherapy triggers exosomal miR-378a-3p and miR-378d release via the EZH2/STAT3 axis, enhancing drug resistance by activating WNT/NOTCH pathways in breast cancer cells[34]. Our qPCR showed exosomal miR-671-5p and miR-378i elevated in K562/DOX cells but not in patients with treatment-resistant AML. The involvement of these miRNAs in leukemia remains understudied. In prostate cancer, miR-671-5p is known to drive tumor progression and metastasis by engaging the NFIA/CRYAB axis[35]. In colon cancer, miR-671-5p is associated with enhanced cell proliferation, migration, and invasion, correlating with a poorer prognosis[36]. Conversely, in glioblastoma multiforme, miR-671-5p modulates radiosensitivity via STAT3 and reduces tumor migration and cancer stem cell (CSC) characteristics by targeting TRAF2[37]. Given the differential expression patterns of exosomal miR-671-5p in AML patients, we speculate that the role of miR-671-5p may vary in different leukemia patients due to different genetic backgrounds. However, contrary to our findings, studies have found that miR-378i expression is reduced in tumor tissue of colon cancer patients[38], and low concentrations of miR-378i may be associated with higher 5-year OS in liver cancer[39]. This discrepancy may stem from the distinct tumor types and their unique molecular landscapes. Notably, our findings reveal that three of the five most highly expressed genes are part of the miR-378 family, suggesting that this gene family may play a significant role in leukemia drug resistance mechanisms and warrants further investigation.
MiR-92b-5p is a member of the oncogenic miR-17~92 gene cluster, which is elevated in a variety of malignancies, including lymphomas, leukemias, lung cancers, breast cancers, and multiple myelomas[40]. Research has shown that miR-92b-5p is highly expressed in cholangiocarcinoma patients[41] and cervical cancer patients with lymph node metastasis[42]. In gastric cancer, TCGA database analysis found that miR-92b-5p is an independent poor prognostic indicator for gastric cancer, and the prognostic model index “ImmiRSig” composed of miR-92b-5p and eight other miRNA molecules was found to be associated with sorafenib and paclitaxel resistance[43]. Exosomal miR-92b-5p may play different roles in different tumors. Consistent with our study, the expression of exosomal miR-92b-5p is found to be increased in breast cancer patients and is related to the stage[44], but its expression level is decreased in lung cancer patients[45]. Given our findings that exosomal miR-92b-5p levels are elevated in leukemia patients with doxorubicin resistance and that its suppression can restore sensitivity to the drug, we propose that miR-92b-5p could serve as a potential molecular marker for predicting AML chemoresistance and a potential target for therapeutic intervention.
In our study, we identified N4BP1 as a target gene for miR-92b-5p for the first time, a discovery we validated through dual-luciferase reporter assays and transfection studies. N4BP1 is a developmentally expressed protein interactor and monoubiquitylation substrate of NEDD4[46], which is mainly located in the nucleus[47]. Research has indicated that N4BP1 is associated with various miRNAs in cancers, such as miR-151 in prostate cancer, miR-28-5p in ovarian cancer, and miR-151a-5p in lung cancer, playing a role in inhibiting tumor progression[48-50]. However, the precise mechanisms underlying N4BP1’s function have not been extensively elucidated in AML. In non-small cell lung cancer cells, N4BP1 binds to ITCH’s WW domain, preventing the polyubiquitination of p73α and c-Jun, which may inhibit tumor growth and increase chemosensitivity through cell cycle arrest and apoptosis[51]. Additionally, it has been documented that NEDD4 interacts with PTEN through its C2 and HECT domains[52]. In our study, Co-IP experiments substantiated the binding of NEDD4 protein to both N4BP1 and PTEN proteins, findings that are in broad agreement with our experimental outcomes. These interactions suggest a complex regulatory network involving N4BP1, NEDD4, and PTEN, which may have significant implications for AML therapeutic strategies.
NEDD4 is a pivotal player in tumorigenesis, modulating the ubiquitination and degradation of key proteins involved in cell proliferation, apoptosis, cell cycle regulation, migration, invasion, epithelial-mesenchymal transition, CSCs, and drug resistance[53]. NEDD4-1 ubiquitinates PTEN at K13 and K289, triggering both mono- and polyubiquitination. Polyubiquitinated PTEN is earmarked for proteasomal degradation, diminishing cytoplasmic PTEN and potentially fostering tumor development, while monoubiquitinated PTEN relocates to the nucleus, increasing nuclear PTEN levels and reducing cytoplasmic levels[29,54-56]. Our findings indicate that PTEN is predominantly monoubiquitinated in doxorubicin-resistant leukemia cells, with a concomitant rise in nuclear PTEN. Moreover, studies have shown that the nuclear PTEN protein binds to the promoter region of RAD51, leading to radio-resistance by promoting homologous recombination repair in glioblastoma[30] and temozolomide resistance[57]. The RAD51 gene family is pivotal in the cellular response to double-strand DNA breaks, managing replication stress, and therefore maintaining genomic integrity and cellular survival. As an ATPase, RAD51 is instrumental in the search for and binding to homologous DNA sequences, thereby facilitating precise and efficient DNA repair[58]. Experiments have demonstrated that suppression of RAD51 expression leads to enhanced DNA damage and increased doxorubicin-induced apoptosis[59,60]. Consistent with these studies, we observed that the nuclear PTEN and RAD51 protein increased in K562/DOX cells and in K562 cells when miR-92b-5p was overexpressed. On the other hand, although we did not observe polyubiquitinated PTEN, our western blots results revealed a decrease in both total and cytoplasmic PTEN protein levels in K562/DOX cells and in K562 cells when miR-92b-5p was overexpressed. The decrease in cytoplasmic PTEN may result from the monoubiquitination of PTEN, which regulates PTEN’s translocation between the cytoplasm and the nucleus. Cytoplasmic PTEN, acting as a tumor suppressor, inhibits the PI3K-AKT-mTOR pathway through its phosphatase activity by dephosphorylating PIP3 to PIP2, which in turn suppresses cell proliferation and stimulates cell apoptosis[61-63]. These findings are in line with our experimental results that miR-92b-5p downregulates cytoplasmic PTEN protein and activates the PI3K-AKT-mTOR pathway in leukemia cell lines. Given our study’s results, we believe that RAD51 and the PI3K-AKT-mTOR pathway are key downstream effectors of miR-92b-5p that are implicated in mediating doxorubicin resistance in leukemia. However, the underlying mechanism for the reduction in total PTEN protein levels remains elusive. The exact pathway through which miR-92b-5p influences total PTEN levels warrants further investigation, including the exploration of post-transcriptional regulation and protein stability.
There are some limitations of this study. The clinical sample size in this study is small, which limits our ability to establish a robust correlation between exosomal miR-92b-5p expression levels and chemoresistance. To address this, we are committed to expanding our clinical sample collection to reduce the influence of random variability and to strengthen the validity of our findings. Additionally, our study focused on the K562 and K562/DOX cell lines, which, while informative, represent a narrow scope of the broader cellular landscape. In future research, we plan to develop a more diverse array of drug-resistant cell lines to provide a comprehensive understanding of the underlying mechanisms. Furthermore, our current research is confined to bioinformatics analysis and in vitro experiments, which, while valuable, do not fully capture the complexity of drug resistance in a clinical context. To bridge this gap, we will incorporate animal models in our future studies to explore the clinical relevance and therapeutic potential of our findings. Lastly, our study did not include the knockdown of NEDD4 and N4BP1, which are critical components in the PTEN mono-ubiquitination pathway. In subsequent studies, we will perform knockdown experiments to elucidate the roles of NEDD4 and N4BP1 in the context of PTEN regulation and doxorubicin resistance. This additional research will provide a more nuanced view of the molecular interactions and contribute to the development of targeted therapies for AML.
In conclusion, we consider that exosomal miR-92b-5p may be a potential molecular marker for predicting AML chemoresistance and a potential target for therapeutic intervention. Our study has demonstrated that miR-92b-5p plays a pivotal role in the regulation of PTEN mono-ubiquitination by suppressing the expression of N4BP1 mRNA and concurrently upregulating the expression of NEDD4 protein. This intricate molecular interplay leads to the nuclear translocation of PTEN, which in turn enhances DNA damage repair mechanisms by upregulating RAD51 and activates the PI3K-AKT-mTOR signaling pathway, thereby inducing doxorubicin resistance in AML. These discoveries shed new light on the resistance mechanisms in AML and offer a promising therapeutic target for clinical intervention.
DECLARATIONS
Acknowledgments
The authors acknowledge the use of Figdraw in the creation of the image presented in Graphical Abstract.
Authors’ contributions
Conceptualization and design: Yao C, Xiao S, Li Q
Experiment and statistical analysis: Li Q, Cheng J
Manuscript drafting: Li Q
Manuscript revision: Qin D, Yao C
All authors contributed to and approved the article.
Availability of data and materials
The data used in this study were obtained from our laboratory experiments. The datasets generated and analyzed during the current study are available from the corresponding author upon reasonable request.
Financial support and sponsorship
This research was supported by a grant from the Key Research and Development program of Hainan Province (ZDYF2020135) and Changsha Natural Science Foundation (kq2202427).
Conflicts of interest
All authors declared that there are no conflicts of interest.
Ethical approval and consent to participate
This study involving human participants was approved by the Ethics Committee of the Third Xiangya Hospital, Central South University (No.23447). Written informed consent was obtained from each participant enrolled in the study.
Consent for publication
Not applicable.
Copyright
© The Author(s) 2025.
Supplementary Materials
REFERENCES
1. Hackl H, Astanina K, Wieser R. Molecular and genetic alterations associated with therapy resistance and relapse of acute myeloid leukemia. J Hematol Oncol. 2017;10:51.
2. He B, Zhao Z, Cai Q, et al. miRNA-based biomarkers, therapies, and resistance in cancer. Int J Biol Sci. 2020;16:2628-47.
3. Giovannetti E, Erozenci A, Smit J, Danesi R, Peters GJ. Molecular mechanisms underlying the role of microRNAs (miRNAs) in anticancer drug resistance and implications for clinical practice. Crit Rev Oncol Hematol. 2012;81:103-22.
4. Bhat AA, Younes SN, Raza SS, et al. Role of non-coding RNA networks in leukemia progression, metastasis and drug resistance. Mol Cancer. 2020;19:57.
5. Seca H, Lima RT, Almeida GM, et al. Effect of miR-128 in DNA damage of HL-60 acute myeloid leukemia cells. Curr Pharm Biotechnol. 2014;15:492-502.
6. Yamamoto H, Lu J, Oba S, et al. miR-133 regulates Evi1 expression in AML cells as a potential therapeutic target. Sci Rep. 2016;6:19204.
7. Liu H, Liu M, Zhang J, Liang Y. Downregulated miR-130a enhances the sensitivity of acute myeloid leukemia cells to adriamycin. Mol Med Rep. 2020;22:2810-6.
8. Pathan M, Fonseka P, Chitti SV, et al. Vesiclepedia 2019: a compendium of RNA, proteins, lipids and metabolites in extracellular vesicles. Nucleic Acids Res. 2019;47:D516-9.
9. Fontana F, Carollo E, Melling GE, Carter DRF. Extracellular vesicles: emerging modulators of cancer drug resistance. Cancers. 2021;13:749.
10. Cariello M, Squilla A, Piacente M, Venutolo G, Fasano A. Drug resistance: the role of exosomal miRNA in the microenvironment of hematopoietic tumors. Molecules. 2022;28:116.
11. Guo QR, Wang H, Yan YD, et al. The role of exosomal microRNA in cancer drug resistance. Front Oncol. 2020;10:472.
12. Wu J, Zhang Y, Li X, et al. Exosomes from bone marrow mesenchymal stem cells decrease chemosensitivity of acute myeloid leukemia cells via delivering miR-10a. Biochem Biophys Res Commun. 2022;622:149-56.
13. Li H, Xie C, Lu Y, Chang K, Guan F, Li X. Exosomal miR92a promotes cytarabine resistance in myelodysplastic syndromes by activating Wnt/β-catenin signal pathway. Biomolecules. 2022;12:1448.
14. Jiang Y, Xiao S, Huang S, et al. Extracellular vesicle-mediated regulation of imatinib resistance in chronic myeloid leukemia via the miR-629-5p/SENP2/PI3K/AKT/mTOR axis. Hematology. 2024;29:2379597.
15. Min QH, Wang XZ, Zhang J, et al. Exosomes derived from imatinib-resistant chronic myeloid leukemia cells mediate a horizontal transfer of drug-resistant trait by delivering miR-365. Exp Cell Res. 2018;362:386-93.
16. Bouvy C, Wannez A, Laloy J, Chatelain C, Dogné JM. Transfer of multidrug resistance among acute myeloid leukemia cells via extracellular vesicles and their microRNA cargo. Leuk Res. 2017;62:70-6.
17. Fishman A, T Lamm A. QsRNA-seq: a protocol for generating libraries for high-throughput sequencing of small RNAs. Bio Protoc. 2019;9:e3179.
18. Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139-40.
19. Betel D, Wilson M, Gabow A, Marks DS, Sander C. The microRNA.org resource: targets and expression. Nucleic Acids Res. 2008;36:D149-53.
20. Rehmsmeier M, Steffen P, Hochsmann M, Giegerich R. Fast and effective prediction of microRNA/target duplexes. RNA. 2004;10:1507-17.
21. Deng YY, Li JQ, Wu SF, et al. Integrated nr database in protein annotation system and its localization. Comput Eng. 2006;32:71-2.
22. Apweiler R, Bairoch A, Wu CH, et al. UniProt: the Universal Protein knowledgebase. Nucleic Acids Res. 2004;32:D115-9.
23. Ashburner M, Ball CA, Blake JA, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. 2000;25:25-9.
24. Tatusov RL, Galperin MY, Natale DA, Koonin EV. The COG database: a tool for genome-scale analysis of protein functions and evolution. Nucleic Acids Res. 2000;28:33-6.
25. Kanehisa M, Goto S, Kawashima S, Okuno Y, Hattori M. The KEGG resource for deciphering the genome. Nucleic Acids Res. 2004;32:D277-80.
26. Koonin EV, Fedorova ND, Jackson JD, et al. A comprehensive evolutionary classification of proteins encoded in complete eukaryotic genomes. Genome Biol. 2004;5:R7.
28. Mao X, Cai T, Olyarchuk JG, Wei L. Automated genome annotation and pathway identification using the KEGG Orthology (KO) as a controlled vocabulary. Bioinformatics. 2005;21:3787-93.
29. Trotman LC, Wang X, Alimonti A, et al. Ubiquitination regulates PTEN nuclear import and tumor suppression. Cell. 2007;128:141-56.
30. Pei X, Mladenov E, Soni A, Li F, Stuschke M, Iliakis G. PTEN loss enhances error-prone DSB processing and tumor cell radiosensitivity by suppressing RAD51 expression and homologous recombination. Int J Mol Sci. 2022;23:12876.
31. Naci D, Berrazouane S, Barabé F, Aoudjit F. Cell adhesion to collagen promotes leukemia resistance to doxorubicin by reducing DNA damage through the inhibition of Rac1 activation. Sci Rep. 2019;9:19455.
32. Fadeev RS, Solovieva ME, Slyadovskiy DA, et al. Cell aggregation increases drug resistance of acute myeloid leukemia cells. Biochem Moscow Suppl Ser A. 2015;9:135-43.
33. Wu T, Chen Y, Du Y, Tao J, Zhou Z, Yang Z. Serum exosomal miR-92b-5p as a potential biomarker for acute heart failure caused by dilated cardiomyopathy. Cell Physiol Biochem. 2018;46:1939-50.
34. Yang Q, Zhao S, Shi Z, et al. Chemotherapy-elicited exosomal miR-378a-3p and miR-378d promote breast cancer stemness and chemoresistance via the activation of EZH2/STAT3 signaling. J Exp Clin Cancer Res. 2021;40:120.
35. Zhu Z, Luo L, Xiang Q, et al. MiRNA-671-5p promotes prostate cancer development and metastasis by targeting NFIA/CRYAB axis. Cell Death Dis. 2020;11:949.
36. Jin W, Shi J, Liu M. Overexpression of miR-671-5p indicates a poor prognosis in colon cancer and accelerates proliferation, migration, and invasion of colon cancer cells. Onco Targets Ther. 2019;12:6865-73.
37. Lin JC, Kuo CY, Tsai JT, Liu WH. miR-671-5p Inhibition by MSI1 promotes glioblastoma tumorigenesis via radioresistance, tumor motility and cancer stem-like cell properties. Biomedicines. 2021;10:21.
38. Gungormez C, Gumushan Aktas H, Dilsiz N, Borazan E. Novel miRNAs as potential biomarkers in stage II colon cancer: microarray analysis. Mol Biol Rep. 2019;46:4175-83.
39. Sun L, Zhou H, Zhao X, Zhang H, Wang Y, Li G. Small RNA sequencing identified miR-3180 as a potential prognostic biomarker for Chinese hepatocellular carcinoma patients. Front Genet. 2023;14:1102171.
40. Yang L, Zhang X, Liu C, Zhang J, Dong B. MiR-92 family members form a cluster required for notochord tubulogenesis in urochordate ciona savignyi. Genes. 2021;12:406.
41. Høgdall D, O'Rourke CJ, Larsen FO, et al. Whole blood microRNAs capture systemic reprogramming and have diagnostic potential in patients with biliary tract cancer. J Hepatol. 2022;77:1047-58.
42. Barquet-Muñoz SA, Pedroza-Torres A, Perez-Plasencia C, et al. microRNA profile associated with positive lymph node metastasis in early-stage cervical cancer. Curr Oncol. 2022;29:243-54.
43. Xu J, Wen J, Li S, et al. Immune-related nine-microRNA signature for predicting the prognosis of gastric cancer. Front Genet. 2021;12:690598.
44. Kan JY, Shih SL, Yang SF, et al. Exosomal microRNA-92b is a diagnostic biomarker in breast cancer and targets survival-related MTSS1L to promote tumorigenesis. Int J Mol Sci. 2024;25:1295.
45. Kim DH, Park H, Choi YJ, et al. Identification of exosomal microRNA panel as diagnostic and prognostic biomarker for small cell lung cancer. Biomark Res. 2023;11:80.
46. Murillas R, Simms KS, Hatakeyama S, Weissman AM, Kuehn MR. Identification of developmentally expressed proteins that functionally interact with Nedd4 ubiquitin ligase. J Biol Chem. 2002;277:2897-907.
47. Sharma P, Murillas R, Zhang H, Kuehn MR. N4BP1 is a newly identified nucleolar protein that undergoes SUMO-regulated polyubiquitylation and proteasomal turnover at promyelocytic leukemia nuclear bodies. J Cell Sci. 2010;123:1227-34.
48. Chiyomaru T, Yamamura S, Zaman MS, et al. Genistein suppresses prostate cancer growth through inhibition of oncogenic microRNA-151. PLoS One. 2012;7:e43812.
49. Xu J, Jiang N, Shi H, Zhao S, Yao S, Shen H. miR-28-5p promotes the development and progression of ovarian cancer through inhibition of N4BP1. Int J Oncol. 2017;50:1383-91.
50. Hu H, Jiang L, Kang X, Zhang L, Zeng S. Extracellular vesicles derived from lung cancer cells promote the progression of lung cancer by delivering miR-151a-5p. Exp Cell Res. 2023;425:113526.
51. Oberst A, Malatesta M, Aqeilan RI, et al. The Nedd4-binding partner 1 (N4BP1) protein is an inhibitor of the E3 ligase Itch. Proc Natl Acad Sci U S A. 2007;104:11280-5.
52. Wang X, Shi Y, Wang J, Huang G, Jiang X. Crucial role of the C-terminus of PTEN in antagonizing NEDD4-1-mediated PTEN ubiquitination and degradation. Biochem J. 2008;414:221-9.
53. Wang ZW, Hu X, Ye M, Lin M, Chu M, Shen X. NEDD4 E3 ligase: functions and mechanism in human cancer. Semin Cancer Biol. 2020;67:92-101.
54. Fouladkou F, Landry T, Kawabe H, et al. The ubiquitin ligase Nedd4-1 is dispensable for the regulation of PTEN stability and localization. Proc Natl Acad Sci U S A. 2008;105:8585-90.
55. Wang X, Trotman LC, Koppie T, et al. NEDD4-1 is a proto-oncogenic ubiquitin ligase for PTEN. Cell. 2007;128:129-39.
56. Drinjakovic J, Jung H, Campbell DS, Strochlic L, Dwivedy A, Holt CE. E3 ligase Nedd4 promotes axon branching by downregulating PTEN. Neuron. 2010;65:341-57.
57. Dong L, Li Y, Liu L, et al. Smurf1 suppression enhances temozolomide chemosensitivity in glioblastoma by facilitating PTEN nuclear translocation. Cells. 2022;11:3302.
58. Bonilla B, Hengel SR, Grundy MK, Bernstein KA. RAD51 gene family structure and function. Annu Rev Genet. 2020;54:25-46.
59. Xie C, Drenberg C, Edwards H, et al. Panobinostat enhances cytarabine and daunorubicin sensitivities in AML cells through suppressing the expression of BRCA1, CHK1, and Rad51. PLoS One. 2013;8:e79106.
60. Lai TH, Ewald B, Zecevic A, et al. HDAC inhibition induces microRNA-182, which targets RAD51 and impairs HR repair to sensitize cells to sapacitabine in acute myelogenous leukemia. Clin Cancer Res. 2016;22:3537-49.
61. Huang ZJ, Zhu JJ, Yang XY, Biskup E. NEDD4 promotes cell growth and migration via PTEN/PI3K/AKT signaling in hepatocellular carcinoma. Oncol Lett. 2017;14:2649-56.
62. Wen W, Li J, Wang L, et al. Inhibition of NEDD4 inhibits cell growth and invasion and induces cell apoptosis in bladder cancer cells. Cell Cycle. 2017;16:1509-14.
Cite This Article
How to Cite
Download Citation
Export Citation File:
Type of Import
Tips on Downloading Citation
Citation Manager File Format
Type of Import
Direct Import: When the Direct Import option is selected (the default state), a dialogue box will give you the option to Save or Open the downloaded citation data. Choosing Open will either launch your citation manager or give you a choice of applications with which to use the metadata. The Save option saves the file locally for later use.
Indirect Import: When the Indirect Import option is selected, the metadata is displayed and may be copied and pasted as needed.
About This Article
Copyright
Data & Comments
Data



























Comments
Comments must be written in English. Spam, offensive content, impersonation, and private information will not be permitted. If any comment is reported and identified as inappropriate content by OAE staff, the comment will be removed without notice. If you have any queries or need any help, please contact us at [email protected].