Galacto-oligosaccharides alone and combined with lactoferrin impact the Kenyan infant gut microbiota and epithelial barrier integrity during iron supplementation in vitro
Abstract
Aim: Iron supplementation to African weaning infants was associated with increased enteropathogen levels. While cohort studies demonstrated that specific prebiotics inhibit enteropathogens during iron supplementation, their mechanisms remain elusive. Here, we investigated the in vitro impact of galacto-oligosaccharides (GOS) and iron-sequestering bovine lactoferrin (bLF) alone and combined on the gut microbiota of Kenyan infants during low-dose iron supplementation.
Methods: Different doses of iron, GOS, and bLF were first screened during batch fermentations (n = 3), and the effect of these factors was studied on microbiota community structure and activity in the new Kenyan infant continuous intestinal PolyFermS model. The impact of different fermentation treatments on barrier integrity, enterotoxigenic Escherichia coli (ETEC) infection, and inflammatory response was assessed using a transwell co-culture of epithelial and immune cells.
Results: A dose-dependent increase in short-chain fatty acid (SCFA) production, Bifidobacterium and Lactobacillus/Leuconostoc/Pediococcus (LLP) growth was detected with GOS alone and combined with bLF during iron supplementation in batches. This was confirmed in the continuous PolyFermS model, which also showed a treatment-induced inhibition of opportunistic pathogens C. difficile and C. perfringens. In all tests, supplementation of iron alone and combined with bLF did not have a significant effect on microbiota composition and activity. We observed a strengthening of the epithelial barrier and a decrease in cell death and pro-inflammatory response during ETEC infection with microbiota fermentation supernatants from iron + GOS, iron + bLF, and iron + GOS + bLF treatments compared to iron alone.
Conclusion: Overall, beneficial effects on infant gut microbiota were shown using advanced in vitro models for GOS alone and combined with bLF during low-dose iron supplementation.
Keywords
INTRODUCTION
Iron deficiency anemia (IDA) is prevalent among African children under 5 years of age[1], and high IDA prevalences of 62% to 73% were reported in Kenyan infants aged 6.5 to 12 months[2,3]. In-home fortification with complementary foods supplemented with iron-containing micronutrient powders (MNPs) is used to reduce IDA in African infants during weaning[4,5]. However, the safety of MNPs containing the WHO-recommended iron dose (10-12.5 mg/day for 6- to 23-month-old infants[6]) was questioned following reports about possible adverse effects on the infant gut microbiota[7]. Iron-induced gut microbial dysbiosis was previously reported for Kenyan and Pakistani infants and characterized by an increase of potential harmful bacteria (Enterobacteriaceae, pathogenic E. coli) and a decrease in beneficial taxa (Bifidobacterium and Lactobacillus)[3,8-10].
Modified iron-containing MNPs with lower iron doses and compounds promoting beneficial gut microbes have been investigated to improve their safety and efficacy. The combination of iron with prebiotics galacto-oligosaccharides (GOS)[3] or with a combination of GOS and fructo-oligosaccharides (FOS)[11] resulted in improved iron absorption and positive effects on the gut microbiota composition in Kenyan infants during weaning. In both studies, the abundances of beneficial Bifidobacterium or Lactobacillus were higher, and the abundances of pathogens and toxin-encoding genes were lower in the feces of infants receiving prebiotics and iron compared to the iron group. The addition of the iron-binding protein bovine lactoferrin (bLF) has been suggested to further improve the safety of iron-containing MNPs. bLF is highly abundant in the whey protein fraction of milk[12] and can promote iron absorption[13]. Other beneficial functions of lactoferrin include its bifidogenic[14], antimicrobial[15], and anti-inflammatory activities[16]. Formula fortified with bLF resulted in a significantly lower incidence of diarrhea in Chinese weaning infants[17] and a lower prevalence of acute gastrointestinal symptoms in young Japanese children[18]. Additionally, bLF improved the iron absorption from a maize-based porridge containing FeSO4 and iron-free LF in Kenyan infants[19]. The efficacy of iron-containing MNPs with the prebiotic GOS and bLF to prevent IDA and microbial dysbiosis is investigated in Kenyan infants[20].
Investigating MNP treatment effects on the gut microbiota in fecal samples does not enable a reliable assessment of the functional impact of the treatments, because intestinal-produced metabolites are largely absorbed and cannot be evaluated in feces[21]. In vitro gut microbiota models circumvent this limitation by investigating the gut microbiota independent of the host and under controlled conditions[22]. For example, the continuous fermentation model PolyFermS inoculated with immobilized fecal microbiota can test several treatments in parallel with the same complex human gut microbiota[23,24], and can be combined with cellular models to study the host-microbe interactions[25]. The PolyFermS model was recently adapted to closely mimic the gut microbiota of Kenyan infants during weaning[26]. A strong prebiotic potential was confirmed for a short-chain GOS/long-chain FOS mixture and inulin but not for acacia gum in Kenyan infant gut microbiota during iron supplementation[27].
The aim of this study was to investigate the direct effects of GOS and bLF, alone or combined, on both the microbe-microbe and host-microbe interactions in the gut microbiota of Kenyan infants during iron supplementation in vitro. First, the effect of different doses of iron as ferrous sulfate, GOS and bLF on the microbiota metabolic activity and growth of infant-characteristic bacteria was assessed during short-term batch fermentations inoculated with artificial microbiota produced from three different Kenyan infant PolyFermS models. Next, the effects of iron, GOS, bLF, and a combination thereof, each at concentrations selected to mimic a double-blind intervention study in Africa[20] on microbiota community structure were investigated in continuous PolyFermS models inoculated with two different infant fecal microbiota. Finally, a transwell co-culture system of epithelial and immune cells was exposed to treated PolyFermS microbiota supernatants to investigate treatment effects on epithelial barrier integrity, pathogen infection and inflammatory response.
METHODS
Fecal donor characteristics and fecal sample collection
Fresh fecal samples from 5 infants aged between 5.6 and 9.7 months living in rural Kenya (Msambweni County) were collected, transported under protective anaerobic and cold conditions, and processed as previously described within less than 30 h for immobilization and inoculation of PolyFermS models[26]. None of the infants received antibiotics prior to sample donation and detailed donor information is given in Supplementary Table 1.
In vitro gut microbiota fermentation experiments
The Kenyan infant PolyFermS model was used to continuously cultivate the donor fecal microbiota as previously described[26]. First, three PolyFermS Kenyan infant fecal microbiota (donor 1, 2 and 3) were used separately to assess the dose-dependent microbiota response to iron, GOS, and bLF during 24 h of batch fermentations in 24-well plates [Figure 1]. Next, the PolyFermS model was used to assess the effect of iron, GOS, and bLF on the microbiota community structure of two Kenyan infant fecal microbiota (donors 4 and 5) [Figure 1].
Figure 1. Overview of batch fermentation and PolyFermS experiments. IR with immobilized Kenyan infant fecal microbiota and connected PolyFermS second-stage treatment reactors (TR1-6). Each fecal microbiota was immobilized and cultivated separately. (A) IR microbiota of donors 1 to 3 were used for batch fermentations in 24-well plates, and (B) IR of donors 4 and 5 were connected to TRs. IR: Inoculum reactors; TRs: test reactors; qPCR: quantitative PCR; HPLC: high-performance liquid chromatography; T: temperature; RT: retention time; STAB: stabilization period; TREAT: treatment period.
Cultivation medium
The cultivation medium was designed to mimic the ileal chyme entering the proximal colon of Kenyan infants during weaning at the age of 6 to 8 months and prepared as previously described[26] and detailed in Supplementary Table 2. For batch fermentations, the medium composition was slightly adapted and contained a 6-fold higher concentration of sodium bicarbonate to enhance the internal buffering capacity. Additionally, carbohydrate sources were reduced by 50% to prevent over-acidification. Further, since phosphate has a strong capacity to bind iron and may affect iron solubility at high concentrations, potassium phosphate monobasic was substituted with 2-morpholinoethanesulfonic acid monohydrate (MES).
Fecal microbiota immobilization and PolyFermS model operation
Immobilization of fecal microbiota and PolyFermS bioreactor operation were performed as reported previously[26] and detailed in Supplementary Materials. Each fecal microbiota was immobilized and cultivated in separate bioreactors, with details provided in Supplementary Materials. After bead colonization and stabilization for at least 21 days, the effluent from the bioreactor with immobilized fecal microbiota served as a microbial inoculum source for batch fermentations. The inoculum reactors (IR) of donors 4 and 5 were connected to second-stage test reactors (TRs) after stabilization, as reported previously[27]. TRs were continuously inoculated with IR effluent (1.25 mL/h) and simultaneously fed with fresh cultivation medium (23.75 mL/h). The operation conditions of IR and TRs were the same, except for stirring, which was 120 rpm for IR and 180 rpm for TR [Figure 1]. TRs were operated for at least 7 days after connection and prior to treatment start, to reach a pseudo-steady state monitored through a day-to-day variation in short-chain fatty acid (SCFA) production of less than 10%[28].
Effluent samples of IR and TRs were collected daily for metabolite analysis with high-performance liquid chromatography (HPLC). The samples (2 mL) were centrifuged for 10 min at 18,407 g and 4 °C. Supernatants were used for HPLC analyses and pellets stored at -80 °C for DNA extraction.
Batch fermentation and PolyFermS experimental setup
Iron (FeSO4·7H2O; Sigma-Aldrich), GOS (Vivinal, 70% GOS, 24% lactose, 6% glucose + galactose; Friesland Campina), and iron-free apo-bLF (Vitalarmor Lactoferrin, Armor Protéines) were used. The treatments were set to mimic an infant's daily oral dose of 5 mg elemental iron, 10 g of Vivinal GOS, and 1 g of apo-bLF, respectively[20]. Therefore, 24.9 mg FeSO4·7H2O/L medium (5 mg elemental iron/L), 11.1 g Vivinal GOS/L medium, and 1.1 g apo-bLF/L medium were used for the 1× treatments in 24-well plates and for the PolyFermS experiments. Dosage calculations considered an estimated infant proximal colon capacity of
Batch fermentations were performed over three consecutive days using fresh effluent microbiota derived from the IR of donors 1, 2, and 3 to screen different treatment doses [Figure 1A]. Iron and bLF were added to the medium at concentrations simulating the in vivo (1×) dose as described above, and half (0.5×) and double (2×) concentrations. In contrast, the Vivinal GOS dose effect was tested at lower levels of 0.12×,
The effect of 1× dose treatments was assessed using PolyFermS model TRs of donors 4 and 5 in two and three repetitions, respectively [Figure 1B]. FeSO4·7H2O was added to the medium components prior to dissolution in dH2O. Vivinal GOS and bLF were dissolved in dH2O separately, filter-sterilized (0.2 µm) and supplemented to the cultivation medium after autoclaving. The iron concentration was measured with Inductively Coupled Plasma - Mass Spectrometry iCap, KED mode in the non-supplemented (2.26 ±
Molecular analysis
The FastDNA SPIN Kit for Soil (MP Biomedicals) was used to extract the DNA of fecal (200 mg) and effluent (pellet of 2 mL) samples according to the manufacturer’s instructions.
Quantitative PCR (qPCR) was performed to determine the absolute numbers of total and selected bacterial targets of the infant gut microbiota (primer details in Supplementary Table 3). Reactions were performed using the Roche Light Cycler 480 System (Hoffmann-La Roche) as previously described[26]. The qPCR gene copy number was adjusted for the median number of 16S rRNA gene copies of each target using the Ribosomal RNA Database[34] to convert the data into absolute bacterial concentrations.
The Illumina MiSeq platform was used to perform paired-end 16S rRNA gene amplicon sequencing (Illumina) at the Genetic Diversity Center (GDC, ETH Zurich) as previously described[26]. The V4 region of the 16S rRNA gene was amplified with the primer combination nxt_515F/nxt_806R (5’-GTGCCAGCMGCCGCGGTAA-3’, 5’-GGACTACHVGGGTWTCTAAT-3’) followed by amplicon barcoding using Nextera Index primers. The DADA2-pipeline[35,36] was used to generate amplicon sequencing variants (ASV) as previously described[26]. Forward and reverse reads were truncated after 170 nucleotides and 160 nucleotides, respectively. Truncated reads with an expected error rate higher than three for forward and four for reverse reads were removed. After filtering, denoising, error rate learning, and ASV inference, reads were merged with a minimum overlap of 40 bp. Chimeric sequences were removed, and taxonomy was assigned using the SILVA database (v.132)[37].
Metabolite analysis
SCFA (acetate, propionate, butyrate, and valerate), branched-chain fatty acids (BCFA, isobutyrate, isovalerate), and intermediate metabolites (succinate, lactate, and formate) were quantified in fecal (200 mg) and effluent (2 mL) samples using HPLC as previously described[26].
Mammalian cell model for microbe-host experiments
To mimic the gut mucosal environment, a transwell plate system was used to co-culture intestinal epithelial Caco-2 (DSMZ ACC 169) and mucin-producing HT29-MTX (ECACC 12040401) cells in the apical and THP-1 Blue cells (Invivogen, thp-nfkb) in the basolateral compartment. THP-1 Blue cells were transfected with a nuclear factor kappa B (NFκB)-inducible secreted embryonic alkaline phosphatase (SEAP) reporter construct (Invivogen), which enables the screening of pro-inflammatory NFκB activation. For simulating pathogen infection, enterotoxigenic Escherichia coli (ETEC) was chosen due to its high prevalence in Kenyan infants (21%-49.2%) and other countries of sub-Saharan Africa[3,9,38].
Bacterial strain and culture conditions
ETEC strain H10407 (ATCC 35401, LGC Standards GmbH) was routinely grown in fresh Luria-Bertani (LB) broth Miller (Becton Dickinson AG) at 37 °C and shaken at 120 rpm (Adolf Kühner AG). ETEC was grown to an optical density of 0.5 (approximately 1.4 × 108 CFU/mL), centrifuged (10 min at 18,407 g), and washed once in phosphate-buffered saline (PBS, Thermo Fisher Scientific) prior to resuspension in minimum essential medium (MEM) Hanks’ Balanced Salts (Thermo Fisher Scientific) for infection experiments.
Co-culture model of epithelial and immune cells
Caco-2 and HT29-MTX cells were cultivated as described in Supplementary Materials. Cells were seeded on cell insert membranes (0.4 µm) of a Millicell 24-well cell culture insert plate (Sigma-Aldrich) in a 75:25 ratio[25] at a final concentration of 5.0 × 104 cells/cm2 and cultivated for 16 to 21 days to reach full differentiation. The medium was exchanged every 2 days in the apical and basolateral compartments of the transwell plate. THP-1 Blue cells were seeded (in RPMI 1640 HEPES medium without antibiotics) into the basolateral compartment at a final concentration of 8.0 × 104 cells/well 24 h prior to combination with the apical insert plate containing the fully differentiated Caco-2/HT29-MTX cell monolayer.
Treatment with PolyFermS microbiota supernatants and infection experiments
Caco-2/HT29-MTX cell monolayers were exposed to PolyFermS supernatant from control and treated donor 4 and donor 5 microbiota (last treatment day of period 1 and 2, respectively) to assess its impact on barrier integrity and infected cell model with ETEC [Figure 2].
Figure 2. Overview of mammalian cell model setup using a transwell system to assess the impact of differently treated PolyFermS supernatant on the epithelial barrier, pathogen infection, and inflammatory response. TEER: Transepithelial electrical resistance; MOI: multiplicity of infection; LDH: lactate dehydrogenase; ETEC: enterotoxigenic Escherichia coli; NFκB: nuclear factor kappa B; SEAP: secreted embryonic alkaline phosphatase.
Effluent samples were centrifuged (15 min at 18,407 g, 4 °C) and the supernatant was filter-sterilized
Statistical analysis and data visualization
Microbiota community analysis was done in R (version 4.0.4) using the phyloseq[39], vegan[40], and ggplot2[41] packages. Differential relative abundance analysis was performed using DESeq2[42]. Rarefied data were used to calculate relative abundances, as well as alpha and beta diversity. GraphPad Prism (v 9.1.0) was used to create graphs and for statistical analysis. Normal distribution was assessed using the Shapiro-Wilk test. A paired t-test was applied to test differences between two independent normal-distributed samples. Welch’s test was used in case of unequal variances and the Mann-Whitney test was applied for samples that were not normally distributed. Differences between more than two independent normally distributed samples were tested with one-way ANOVA, and in case of statistical significance, a Dunnet’s post-hoc test was performed. The Kruskal-Wallis test was used for samples that were not normally distributed with a post hoc Dunn’s test in case of statistical significance. The statistical level of significance was set to P < 0.05.
RESULTS
Dose-dependent microbiota response to iron, GOS, and bLF supplementation during batch fermentation
The response to different doses of iron, GOS, and bLF was assessed during 24 h batch fermentations in 24-well plates inoculated with three different Kenyan infant PolyFermS microbiota [Supplementary Figure 2], and key infant gut bacterial taxa growth and metabolite production were evaluated.
Iron supplementation with GOS, alone or combined with bLF, stimulated Bifidobacterium and Lactobacillus/Leuconostoc/Pediococcus (LLP) [Table 1], increased total metabolic activity [Table 2] and consequently decreased the final pH by about 1.5 pH unit at the highest GOS dose [Supplementary Table 4] compared to iron (1×) alone in all three microbiota. GOS response increased with the dose (P < 0.05) for LLP and for total metabolic activity. For microbiota 1, the production of acetate, propionate, and butyrate increased with the GOS level, while for microbiota 2, mainly acetate and formate, and for microbiota 3, acetate, formate, succinate, and lactate responded to GOS [Table 2 and Supplementary Table 6]. Growth of potential pathogenic taxa Clostridioides difficile, Clostridium perfringens, and Enteropathogenic Escherichia coli were not affected by the treatments in all three microbiota [Supplementary Table 5].
Quantification of key bacterial taxa in Kenyan infant PolyFermS microbiota treated with iron, GOS and bLF during 24 h batch fermentations in 24-well plates
| Bifidobacterium (bacteria/mL) | LLP (bacteria/mL) | ||||||
| Donor 1 | Donor 2 | Donor 3 | Donor 1 | Donor 2 | Donor 3 | ||
| log10 | Inoculum | 9.70 ± 0.31 | 8.99 ± 0.08 | ND | 9.10 ± 0.08 | 8.73 ± 0.36 | ND |
| Control | 7.63 ± 0.23 | 9.30 ± 0.27 | 8.24 ± 0.11 | 7.43 ± 0.06 | 7.89 ± 0.26 | 6.24 ± 0.56 | |
| Δlog10 compared to control | Iron 0.5× | -0.12 ± 0.50 | 0.02 ± 0.03 | -0.09 ± 0.09 | 0.09 ± 0.05 | 0.15 ± 0.30 | 0.16 ± 0.18 |
| Iron 1× | 0.06 ± 0.57 | 0.05 ± 0.09 | 0.00 ± 0.08 | 0.15 ± 0.12 | 0.02 ± 0.07 | 0.29 ± 0.12 | |
| Iron 2× | -0.17 ± 0.42 | -0.19 ± 0.12 | -0.03 ± 0.08 | 0.00 ± 0.11 | -0.14 ± 0.12 | 0.19 ± 0.17 | |
| Iron 1× bLF 0.5× | -0.06 ± 0.28 | 0.11 ± 0.15 | -0.07 ± 0.07 | 0.01 ± 0.11 | 0.01 ± 0.00 | 0.17 ± 0.12 | |
| Iron 1× bLF 1× | 0.05 ± 0.48 | 0.06 ± 0.06 | -0.10 ± 0.13 | 0.19 ± 0.05 | 0.08 ± 0.05 | 0.22 ± 0.27 | |
| Iron 1× bLF 2× | -0.11 ± 0.55 | -0.09 ± 0.07 | -0.09 ± 0.13 | 0.08 ± 0.19 | 0.01 ± 0.09 | 0.11 ± 0.28 | |
| Iron 1× GOS 0.12× | -0.01 ± 0.10 | 0.16 ± 0.07 | 0.23 ± 0.11 | 0.08 ± 0.06 | -0.07 ± 0.24 | 0.38 ± 0.10 | |
| Iron 1× GOS 0.25× | 0.03 ± 0.46 | 0.43 ± 0.09*** | 0.39 ± 0.17 | 0.26 ± 0.19 | 0.20 ± 0.04 | 0.71 ± 0.23 | |
| Iron 1× GOS 0.5× | 0.43 ± 0.66 | 0.47 ± 0.06*** | 0.59 ± 0.24* | 0.51 ± 0.09* | 0.44 ± 0.15* | 1.11 ± 0.21** | |
| Iron 1× bLF 0.5× GOS 0.12× | 0.07 ± 0.09 | 0.22 ± 0.09 | 0.17 ± 0.11 | 0.01 ± 0.11 | 0.19 ± 0.10 | 0.33 ± 0.14 | |
| Iron 1× bLF 1× GOS 0.25× | 0.13 ± 0.18 | 0.43 ± 0.12** | 0.35 ± 0.19* | 0.20 ± 0.15 | 0.33 ± 0.04** | 0.55 ± 0.04* | |
| Iron 1× bLF 2× GOS 0.5× | 0.33 ± 0.23* | 0.46 ± 0.22** | 0.49 ± 0.22* | 0.29 ± 0.06* | 0.52 ± 0.14*** | 0.74 ± 0.15** | |
Quantification of total metabolites and SCFA in Kenyan infant PolyFermS microbiota treated with iron, GOS and bLF during 24 h batch fermentations in 24-well plates
| Total metabolites (mM) | Acetate (mM) | Propionate (mM) | Butyrate (mM) | Formate (mM) | ||||||||||||
| Donor 1 | Donor 2 | Donor 3 | Donor 1 | Donor 2 | Donor 3 | Donor 1 | Donor 2 | Donor 3 | Donor 1 | Donor 2 | Donor 3 | Donor 1 | Donor 2 | Donor 3 | ||
| mM | Inoculum | 108.39 ± 4.28 | 96.10 ± 4.06 | ND | 57.68 ± 0.60 | 52.62 ± 1.80 | ND | 27.79 ± 4.17 | 27.25 ± 2.55 | ND | 5.39 ± 0.65 | 9.81 ± 0.49 | ND | 16.14 ± 13.96 | 4.25 ± 0.09 | ND |
| Control | 88.24 ± 9.18 | 81.30 ± 6.57 | 86.90 ± 1.33 | 51.88 ± 13.93 | 38.35 ± 4.97 | 49.48 ± 3.32 | 8.98 ± 1.52 | 19.97 ± 1.26 | 4.10 ± 0.56 | 12.20 ± 1.51 | 5.75 ± 1.94 | 6.76 ± 0.39 | 20.91 ± 5.64 | 13.66 ± 1.90 | 18.75 ± 3.94 | |
| ΔmM compared to control | Iron 0.5× | -4.70 ± 5.11 | -0.36 ± 3.87 | -1.24 ± 3.07 | -4.04 ± 2.60 | -1.15 ± 1.83 | -1.78 ± 0.56 | 0.38 ± 0.40 | -0.49 ± 0.46 | -0.18 ± 0.13 | -0.71 ± 0.37 | 0.20 ± 0.51 | -0.94 ± 0.25 | -0.15 ± 1.19 | 0.87 ± 0.33 | 1.03 ± 2.35 |
| Iron 1× | -4.59 ± 1.98 | -1.03 ± 2.41 | -2.09 ± 2.93 | -5.70 ± 4.37 | -1.68 ± 1.56 | -0.76 ± 0.49 | 0.63 ± 1.32 | -0.17 ± 0.31 | -0.15 ± 0.23 | -0.52 ± 0.43 | 0.38 ± 0.66 | -1.06 ± 0.39 | -0.97 ± 1.60 | 0.68 ± 0.94 | -0.50 ± 2.09 | |
| Iron 2× | -2.29 ± 4.23 | 0.11 ± 1.68 | -0.20 ± 2.13 | -2.18 ± 2.70 | -0.56 ± 0.35 | 1.17 ± 0.98 | 1.26 ± 1.12 | 0.46 ± 0.43 | -0.03 ± 0.24 | -0.44 ± 0.93 | 0.69 ± 0.84 | -0.61 ± 0.13 | -0.90 ± 1.61 | -0.17 ± 0.46 | -1.15 ± 1.83 | |
| Iron 1× bLF 0.5× | -6.70 ± 4.62 | -3.26 ± 2.42 | -3.61 ± 2.26 | -5.35 ± 7.98 | -1.72 ± 1.50 | -1.27 ± 0.36 | 0.64 ± 0.75 | -0.52 ± 0.33 | 0.17 ± 0.14 | -1.00 ± 0.08 | -0.49 ± 1.11 | -1.48 ± 0.28 | -0.71 ± 4.59 | -0.24 ± 1.25 | -1.73 ± 1.82 | |
| Iron 1× bLF 1× | -7.34 ± 4.69 | -1.72 ± 2.29 | -2.57 ± 2.10 | -5.36 ± 9.19 | -1.59 ± 1.19 | -1.02 ± 0.46 | 0.99 ± 1.50 | -0.24 ± 0.05 | 0.27 ± 0.10* | -1.42 ± 0.14* | -0.19 ± 0.88 | -1.87 ± 0.31* | -1.24 ± 6.13 | 0.44 ± 1.35 | -1.02 ± 1.73 | |
| Iron 1× bLF 2× | -1.39 ± 4.33 | 3.74 ± 3.96 | -0.16 ± 2.68 | -0.27 ± 6.29 | 1.44 ± 2.79 | 2.93 ± 0.20*** | 1.74 ± 1.13 | 0.96 ± 0.56* | 1.00 ± 0.14**** | -1.58 ± 0.69 | 0.53 ± 1.15 | -2.42 ± 0.34** | -1.36 ± 3.83 | 1.01 ± 1.18 | -3.47 ± 2.05 | |
| Iron 1× GOS 0.12× | 8.24 ± 2.53** | 13.46 ± 2.02 | 8.01 ± 2.38 | 2.30 ± 1.42 | 8.10 ± 1.71 * | -1.58 ± 1.02 | 1.27 ± 0.20 | 1.64 ± 0.35 | -0.31 ± 0.66 | 2.96 ± 0.63 | 1.03 ± 0.38 | -0.39 ± 0.75 | 1.16 ± 1.57 | 2.43 ± 0.58 | 8.68 ± 2.57 | |
| Iron 1× GOS 0.25× | 16.67 ± 4.45*** | 23.95 ± 3.92 | 19.52 ± 4.33** | 2.13 ± 2.51 | 18.41 ± 4.30*** | -0.29 ± 0.46 | 3.74 ± 0.80 | 3.01 ± 1.17 | -1.01 ± 0.43 | 6.05 ± 1.02** | 0.82 ± 0.88 | 0.18 ± 0.77 | 4.10 ± 3.08 | 1.85 ± 2.03 | 16.21 ± 4.06 | |
| Iron 1× GOS 0.5× | 30.11 ± 4.28**** | 45.95 ± 6.20** | 37.59 ± 7.64*** | 5.81 ± 5.90* | 30.42 ± 6.31**** | 3.80 ± 0.14** | 5.48 ± 2.44** | 9.80 ± 2.81* | -2.37 ± 0.61*** | 10.80 ± 3.25*** | 0.36 ± 3.62 | 0.25 ± 0.74 | 7.46 ± 5.76 | 5.83 ± 4.39 | 24.89 ± 6.26** | |
| Iron 1× bLF 0.5× GOS 0.12× | 2.75 ± 0.64* | 7.06 ± 1.32 | 7.50 ± 1.38 | -3.30 ± 3.32 | 4.26 ± 1.94 | -1.79 ± 1.51 | 1.45 ± 0.86 | 1.19 ± 0.84 | -0.04 ± 0.55 | 2.46 ± 1.07 | 0.15 ± 0.72 | -0.71 ± 0.47 | 2.56 ± 1.81 | 1.52 ± 1.31 | 8.07 ± 0.67* | |
| Iron 1× bLF 1× GOS 0.25× | 13.36 ± 2.52**** | 19.91 ± 1.04 | 19.28 ± 3.10 | 0.57 ± 2.69 | 14.89 ± 5.27** | -0.06 ± 1.93 | 2.90 ± 0.12 | 3.48 ± 0.58 | -0.83 ± 0.41 | 5.53 ± 1.50 | 0.53 ± 1.78 | -0.39 ± 0.67 | 4.56 ± 3.32 | 0.99 ± 2.48 | 15.72 ± 2.86*** | |
| Iron 1× bLF 2× GOS 0.5× | 33.93 ± 2.81**** | 42.90 ± 1.18** | 37.18 ± 7.46*** | 9.59 ± 3.69** | 30.90 ± 6.53**** | 7.09 ± 1.77** | 3.96 ± 3.25* | 9.72 ± 2.26* | -2.11 ± 0.53*** | 10.54 ± 2.32** | 0.15 ± 3.76 | 0.36 ± 0.33 | 9.34 ± 10.05 | 2.61 ± 4.68 | 22.50 ± 5.19**** | |
GOS supplementation stimulates SCFA production and growth of beneficial gut microbes while inhibiting potential pathogens during long-term continuous cultivation
Two independent continuous PolyFermS models inoculated with gut microbiota from Kenyan infants 4 and 5 were used to directly test the effect of 1× iron, GOS, and bLF supplementation on the microbiota community structure, dynamics, and metabolite production.
The composition and metabolite profile of the two PolyFermS microbiota after initial colonization and stabilization differed [Supplementary Figures 3 and 4]. PolyFermS microbiota of donor 4 was dominated by Bifidobacterium, Streptococcus, Lactococcus, and Veillonella, which was akin to the fecal microbiota, and produced high concentrations of acetate, propionate, and formate. PolyFermS microbiota of donor 5 was dominated by Bacteroides, Streptococcus, and Megasphaera and produced high concentrations of acetate and butyrate. All parallel reactors of donor 4 or donor 5 PolyFermS microbiota exhibited a similar baseline at the end of the stabilization periods of 7 to 9 days, enabling the evaluation of different treatment combinations on a similar microbiota.
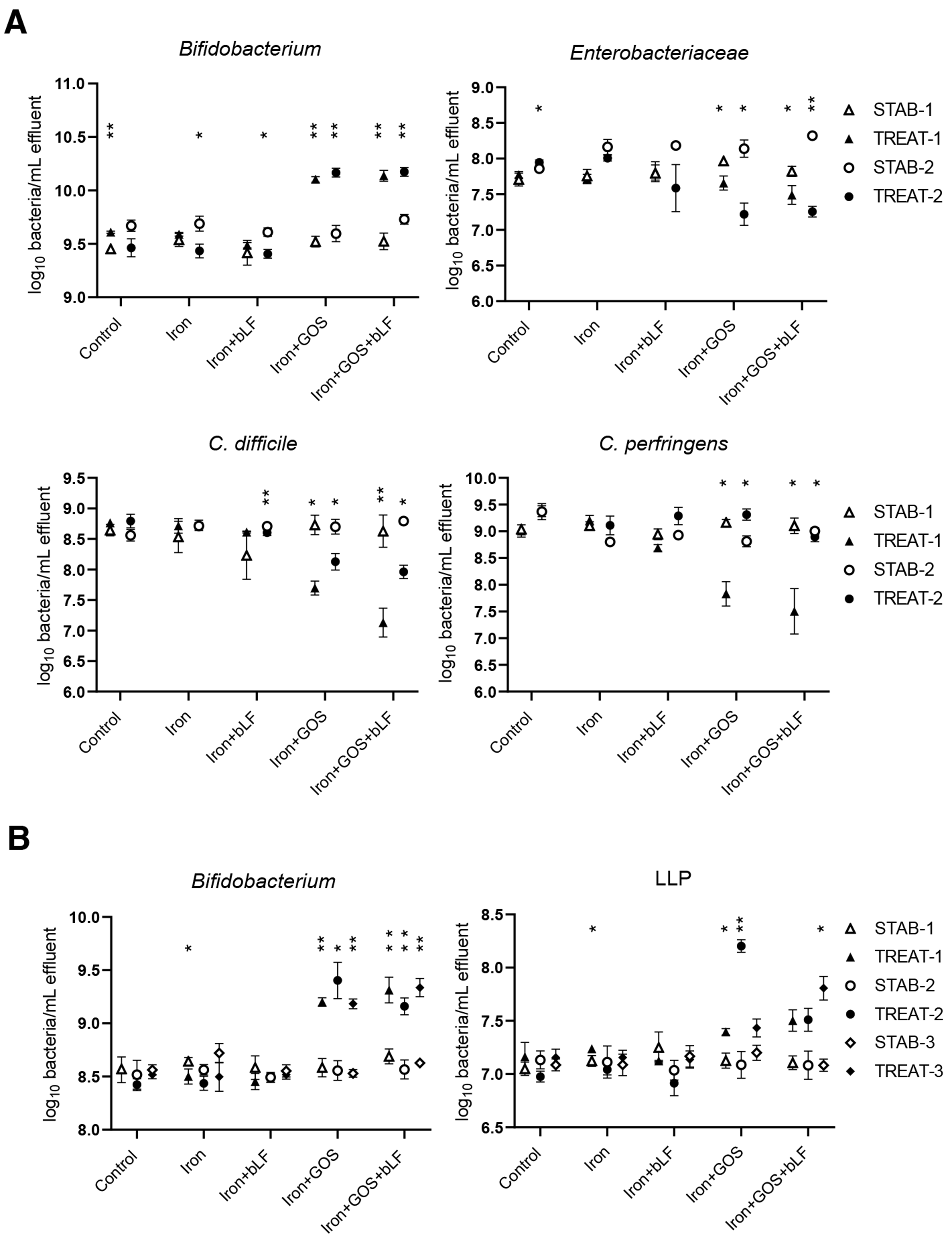
In line with batch experiments, supplementation of GOS during iron treatment stimulated the growth of beneficial bacteria during continuous cultivation [Figure 3]. Iron with GOS alone and combined with bLF resulted in a similar increase in Bifidobacterium at the end of all treatment periods compared to the levels at stabilization for donor 4 (average increase of 0.58 log/mL for GOS and 0.53 log/mL for GOS + bLF) and donor 5 microbiota (average increase of 0.70 log/mL for GOS and 0.64 log/mL for GOS + bLF). LLP showed an increase only in donor 5 microbiota upon GOS (average increase of 0.54 log/mL) and GOS + bLF (average increase of 0.52 log/mL) treatment during iron supplementation compared to stabilization levels. A similar decrease in potentially pathogenic bacteria was detected in donor 4 microbiota with GOS and GOS + bLF for Enterobacteriaceae (average decrease of -0.61 and -0.70 log/mL, respectively) and C. difficile (average decrease of -0.80 and -1.16 log/mL, respectively). Additionally, Clostridium perfringens decreased during treatment period 1 (-1.33 log/mL for GOS and -1.60 log/mL for GOS + bLF), but not during period 2 in donor 4 microbiota. Supplementation with iron alone and iron with bLF did not result in major repeatable changes in bacterial concentrations during treatment compared to the non-supplemental control. In donor 5 microbiota, however, Bifidobacterium decreased during treatment period 1 with iron supplementation (-0.14 log/mL) and C. difficile increased in two of three treatment periods with iron (+0.24 and +1.13 log/mL) and iron + bLF (+0.25 and +0.82 log/mL) supplementation compared to the non-supplemented control (+0.15 and +0.51 log/mL) [Supplementary Figure 5]. Total bacterial counts were not different between stabilization and treatment periods [Supplementary Figure 5].
Figure 3. Quantification of beneficial and potential pathogenic taxa before and after treatment with iron, GOS, and bLF in PolyFermS. Mean ± SD of log10 bacteria/mL effluent is shown for the last three days of stabilization (STAB) and the last three days of treatment (TREAT) of (A) two experimental periods of donor microbiota 4 and (B) three experimental periods of donor microbiota 5. Significant differences between STAB and TREAT in the corresponding periods are indicated. *P < 0.05, **P < 0.01. Enterobacteriaceae and C. difficile were also detected in donor 5 [Supplementary Figure 5]. GOS: Galacto-oligosaccharides; bLF: bovine lactoferrin.
Treatment effects on community composition and structure were assessed by 16S rRNA gene amplicon sequencing. Principal coordinate analysis (PCoA) of weighted Jaccard distance revealed distinct clustering of microbiota treated with GOS alone and GOS + bLF during iron supplementation in donor 4 (driven by Bifidobacterium and Eggerthella, Figure 4A) and donor 5 microbiota (driven by Bifidobacterium and Clostridium sensu stricto 1, Figure 4B). Compared with iron supplementation alone, significant shifts in weighted Jaccard distance were detected for treatments with GOS alone and combined with bLF in donor 4 [Supplementary Figure 6B] and 5 [Supplementary Figure 7B] microbiota.
Figure 4. PCoA of binary and weighted Jaccard distance metrics of before and after treatment with iron, GOS and bLF in PolyFermS. The last three days of stabilization and treatment of two and three experimental periods are shown for (A) donors 4 and (B) 5, respectively. The top 8 genera associated with the community composition are plotted as vectors in weighted Jaccard. PCoA: Principal coordinate analysis; GOS: galacto-oligosaccharides; bLF: bovine lactoferrin.
Differential abundance analysis with DESeq2 detected several genera that were significantly different in relative abundance during the last three days of treatment compared to the last three days of stabilization
Figure 5. Differential abundance analysis (DESeq2) at genus level after treatment with iron and GOS with/without bLF in PolyFermS. Barplots show log2-fold changes of genera significantly (P < 0.05) different in relative abundance between the last three days of iron co-supplementation with GOS and/or bLF and the last three days of supplementation with iron alone for all experimental periods in (A) donor 4 and (B) donor 5 microbiota. GOS: Galacto-oligosaccharides; bLF: bovine lactoferrin.
Co-supplementation of iron + GOS, alone or combined with bLF, resulted in consistent changes in both tested microbiota with increased relative abundance of Bifidobacterium and Eggerthella and decreased Clostridioides (assigned to C. difficile, up to -4.3 log2FoldChange) [Figure 5]. Further, the relative abundance of genus Clostridium sensu stricto decreased in donor 4 (all ASVs were assigned to C. perfringens) while it increased in donor 5 (all ASVs were assigned to C. neonatale) during iron + GOS and iron + GOS + bLF supplementation compared to iron alone supplementation [Figure 5].
No consistent differences in genus relative abundance were detected when comparing the microbiota treated with iron + GOS to those treated with iron + GOS + bLF. Further, the microbiota community evenness decreased for donor 4 microbiota after iron + GOS and iron + GOS + bLF treatment compared to the stabilization period, likely associated with the promotion of the dominant Bifidobacterium leading to a more uneven community [Supplementary Figure 9].
As expected, total metabolite production was increased upon iron + GOS (+58 to 76 mM) and iron + GOS + bLF (+24 to 36 mM) supplementation in both donor microbiota compared to the stabilization period concentrations [Figure 6 and Supplementary Figure 10]. Production of acetate in both donors and propionate in donor 4 and butyrate in donor 5 microbiota was enhanced in treatments containing GOS compared to stabilization.
Figure 6. Quantification of total and intermediate metabolites and SCFA after treatment with iron and GOS with/without bLF in PolyFermS. Mean ± SD of metabolite concentration is shown for the last three days of stabilization (STAB) and the last three days of treatment (TREAT) of (A) two experimental periods of donor 4 and (B) three experimental periods of donor 5. Significant differences between STAB and TREAT in the corresponding periods are indicated. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001. SCFA: Short-chain fatty acid; GOS: galacto-oligosaccharides; bLF: bovine lactoferrin.
In summary, no effect of iron supplementation alone was observed on microbiota composition and metabolic activity. The addition of GOS alone and combined with bLF during iron supplementation promoted SCFA production and beneficial taxa during continuous cultivations in PolyFermS and decreased potential harmful bacteria, including C. difficile and C. perfringens.
GOS and bLF-treated microbiota supernatants strengthen the epithelial barrier and protect from infection-induced effects donor-dependently
Finally, it was assessed whether the fermentation supernatants from treated PolyFermS microbiota with iron, iron + bLF, iron + GOS, and iron + GOS + bLF affected the epithelial barrier, infection, and inflammatory response during ETEC infection differently from the non-treated PolyFermS microbiota in an in vitro model of epithelial and immune cells.
The epithelial barrier was strengthened when all types of PolyFermS supernatants were added, as indicated by higher TEER values compared to the negative control PBS [Figure 7]. The increase in TEER was higher with the supernatant from iron + GOS + bLF compared to iron-alone treated microbiota of donor 4 (1,160 vs. 989 Ω·cm2) [Figure 7A]. ETEC adhesion and invasion were not impacted by the addition of any of the microbiota supernatants [Figure 7] or direct iron supplementation at 5 mg/L [Supplementary Figure 11]. Infection-induced cell death (assessed by LDH release) was only decreased by supernatant of iron + GOS- and iron + GOS + bLF-treated donor 5 microbiota compared to iron-treated microbiota [Figure 7B]. The infection-induced pro-inflammatory response was stimulated when the control microbiota supernatant was added, with a 3.6-fold increased NFκB activation in control compared to PBS [Figure 7]. In contrast, the addition of iron + GOS- and iron + GOS + bLF-treated supernatant of donor 4 microbiota did not result in increased NFκB activation [Figure 7A]. Iron + bLF supernatant of donor 5 microbiota significantly reduced the pro-inflammatory response compared to iron alone supernatant [Figure 7B]. TEER values obtained with donor 4 supernatants were negatively correlated with the relative abundance of Finegoldia (Spearman r =
Figure 7. Effect of PolyFermS supernatant treatments on epithelial barrier, infection and inflammation in a Caco-2/HT29-MTX/THP-1 Blue cell co-culture model. Barrier integrity was assessed by TEER measurement after 24-h exposure to 20% of microbiota supernatant of (A) donor 4 and (B) donor 5. CFU of adhered and invaded ETEC and cell cytotoxicity were assessed by plating and LDH release, respectively, after 3 h of infection in the presence of 20% microbiota supernatant. NFκB activation in THP1-Blue cells was assessed after 21 h of further incubation following infection. Mean ± SD is shown, n = 3 independent cell passages. TEER: Transepithelial electrical resistance; CFU: colony forming units; ETEC: enterotoxigenic Escherichia coli; LDH: lactate dehydrogenase; NFκB: nuclear factor kappa B.
In summary, a strengthening of the epithelial barrier and a decrease in cell death and pro-inflammatory response during ETEC infection was observed with microbiota fermentation supernatants from iron + GOS, iron + bLF, and iron + GOS + bLF treatments compared to iron alone.
DISCUSSION
In this study, we investigated the effect of low-dose iron supplementation, alone and combined with bLF, GOS, and GOS + bLF, on the gut microbiota of infants living in a rural area of Kenya and at the age of weaning (5.6-9.7 months old) in the recently validated Kenyan infant PolyFermS microbiota model[26].
Iron supplementation mimicking 5 mg of iron/day did not induce substantial changes in the five investigated Kenyan infant gut microbiota during batch and continuous fermentations in vitro. This agrees with our previous Kenyan infant PolyFermS study[27], where iron supplementation mimicking a higher dose of 12.5 mg iron/day also did not induce major changes in four investigated Kenyan infant gut microbiota, except for an increase in C. difficile abundance in one microbiota as observed here in donor 5 microbiota as well. Other in vitro studies also reported none[43] or minimal[44] changes in human gut microbiota composition with iron supplementation. These in vitro studies are in contrast to the in vivo observed increase in Enterobacteriaceae or enteropathogens and decrease in Bifidobacterium in the fecal microbiota of 6.0-9.5-month-old Kenyan infants after iron fortification (5 and 12.5 mg iron/day)[3,8,9]. This suggests that the adverse effects of iron on the fecal microbiota reported in vivo may be due to not yet identified iron-host-microbiota interactions in specific individuals rather than being a result of direct iron-gut microbiota interactions.
The addition of GOS alone and combined with bLF during iron supplementation promoted SCFA production and growth of beneficial Bifidobacterium and LLP, while a decrease in potentially harmful bacteria, including C. difficile and C. perfringens was observed. This observation agrees with our previous studies where increased Bifidobacterium and/or Lactobacillus relative abundance concomitant with the decreased enteropathogen markers was observed in the microbiota of weaning 6.0-9.5-month-old Kenyan infants treated with GOS[3] or with a mixture of short-chain GOS (90%) and long-chain FOS (10%) during iron supplementation in vitro[27] or in vivo[11]. The bifidogenic and metabolic effects of GOS have been reported in cohort studies and in vitro fermentation studies with Western infants aged 4 to 24 months[45-50]. Growth inhibition of GOS on enteropathogens in our study may be explained by the production of bacteriocins by GOS-promoted bifidobacteria[51,52] and by the higher SCFA levels, as SCFA were inversely correlated with
The addition of bLF during iron supplementation did not impact the composition and metabolite profile of Kenyan infant gut microbiota in our in vitro study. Previous clinical studies in young pre-weaning infants reported an effect of bLF supplementation, with an increase in Bacteroides and a reduction in Enterobacter, Klebsiella, Staphylococcus, Haemophilus, and Lactobacillus compared to control groups[56-58]. In contrast, a recent study found no impact of bLF on the fecal microbiota and metabolome of 8-month-old infants[59]. The antimicrobial activity of bLF against enteropathogens, such as pathogenic E. coli and Clostridium spp., was observed in mice and in pure culture experiments[60-63]. In our study, the lack of antimicrobial effect of apo-bLF (< 5% iron saturated) on C. difficile and C. perfringens may be partly explained by the low tested dose of 1.1 mg/mL that mimicked the daily dose of 1 g bLF in the cohort trial[20]. It was previously reported that growth inhibition of C. difficile and C. perfringens by bLF (15%-20% iron saturated) occurred at a minimal concentration of 16 mg/mL[61]. Another possible explanation for the lack of pathogen inhibition in our experiments might be the form in which bLF was supplemented in our model. In an in vitro gut microbiota model for C. difficile infection, 5 mg/mL of apo-bLF did not affect C. difficile growth and toxin production while holo-bLF (85% iron saturated) led to inhibition of both[64].
Several studies reported iron-induced adhesion of opportunistic enteropathogens, such as E. coli and Salmonella, to epithelial cells in vitro[25,65,66]. Adhesion of ETEC to HT29 cells doubled when a high dose of 100 µM iron was added but not with a lower iron dose of 50 µM, compared to the control[66]. This iron dose effect on ETEC adhesion may explain our results, as in our ETEC infection study of Caco-2/HT29-MTX cells, iron concentration in the supplemented fermentation supernatants were low with 0.4 mg/L (0.7 µM) and 0.6 mg/L (1.1 µM) for donor 4 and 5, respectively. Direct iron supplementation of cell medium at 5 mg/L (9 µM) to mimic the daily dose of 5 mg iron in the cohort trial[20] also did not impact ETEC infection. Interestingly, infection-induced cell death was decreased by supernatant from donor 5 microbiota treated with bLF and GOS during iron supplementation and this was correlated with increased concentrations of acetate and butyrate in those supernatant samples. Butyrate was previously shown to suppress Caco-2 cell death[67], while acetate and butyrate are energy substrates for epithelial cells[68].
One limitation of this study is that the 16S rRNA gene short-amplicon sequencing approach does not provide taxonomic species- or strain-level resolution. Employing metagenomic sequencing could overcome this limitation, offering detailed insights into whether the predominant fecal Bifidobacterium strains were retained in vitro. Moreover, metagenomic data could elucidate potential cross-feeding interactions in response to GOS-containing treatments, thereby enhancing our mechanistic understanding of these treatments. Another study limitation is that with the used in vitro models, we could not observe an iron-induced microbial dysbiosis as previously reported in vivo. In vitro gut microbiota modeling does not account for all host- and environmental-related factors, and the conditions of the fermentations were tightly set[26]. Combining different microbiota models (in vitro, ex vivo, in silico, and animal) and integrating them with cohort data may help overcome this limitation when studying complex microbial ecosystems in disease[22].
In conclusion, this in vitro study is the first to evaluate the impact of a novel combination of GOS with bLF on Kenyan infant microbiota during low-dose iron supplementation. Using the Kenyan infant PolyFermS microbiota model, we demonstrated that supplementation with GOS alone or combined with bLF elicited bifidogenic effects, inhibited enteropathogens, and promoted SCFA production, while bLF and iron alone showed no significant impact. Additionally, combining in vitro-treated microbiota with a mammalian cell model identified donor-dependent beneficial effects of GOS and bLF on the epithelial barrier and immune response. The results from this in vitro study will aid in interpreting outcomes from a parallel cohort study in the same infant population, advancing our understanding of the potential benefits of GOS combined with bLF in enhancing infant gut health during iron supplementation.
DECLARATIONS
Authors’ contributions
Designed the study: Rachmühl C, Lacroix C, Geirnaert A
Obtained funding: Brittenham GM, Zimmermann MB, Lacroix C
Collected fecal samples: Giorgetti A, Stoffel NU
Maintained cell lines and prepared the co-cultures for experiments: Ferragamo A, Rachmühl C
Performed the experiments and analyzed the data: Rachmühl C
Data interpretation: Rachmühl C, Geirnaert A, Lacroix C
Wrote the manuscript: Rachmühl C, Geirnaert A
Reviewed the manuscript: Lacroix C
All authors read and approved the submitted manuscript.
Availability of data and materials
The datasets generated and/or analyzed during the current study are available in the European Nucleotide Archive (ENA) repository, accession number PRJEB63476 (https://www.ebi.ac.uk/ena/browser/view/ PRJEB63476). Other raw data that support the findings of this study are available from the corresponding author upon reasonable request.
Financial support and sponsorship
This study was funded by the National Institute of Diabetes and Digestive and Kidney Diseases (National Institutes of Health, USA). Grant no. R01 DK115449.
Conflicts of interest
Lacroix C is an Editorial Board member of the journal Microbiome Research Reports. Lacroix C was not involved in any steps of editorial processing, notably including reviewers’ selection, manuscript handling, and decision-making. The other authors declare that there are no conflicts of interest.
Ethical approval and consent to participate
The study was part of the clinical trial “Prebiotic GOS and Lactoferrin for Beneficial Gut Microbiota With Iron Supplements” registered at clinicaltrials.gov as NCT03866837. The Ethics Commission of ETH Zürich, Switzerland (EK 2019-N-04), the Kenya Medical Research Institute (KEMRI) Scientific and Ethics Review Unit (SERU) (KEMRI/RES/7/3/1 no.656), the Columbia University Institutional Review Boards (IRB), USA (IRB-AAAR8900) reviewed and approved this study. Informed consent was obtained from the parents or the legal guardians of the infants. All methods were carried out in accordance with relevant guidelines and regulations.
Consent for publication
Not applicable.
Copyright
© The Author(s) 2024.
Supplementary Materials
REFERENCES
1. Gedfie S, Getawa S, Melku M. Prevalence and associated factors of iron deficiency and iron deficiency anemia among under-5 children: a systematic review and meta-analysis. Glob Pediatr Health 2022;9:2333794X221110860.
2. Giorgetti A, Paganini D, Nyilima S, et al. The effects of 2’-fucosyllactose and lacto-N-neotetraose, galacto-oligosaccharides, and maternal human milk oligosaccharide profile on iron absorption in Kenyan infants. Am J Clin Nutr 2023;117:64-72.
3. Paganini D, Uyoga MA, Kortman GAM, et al. Prebiotic galacto-oligosaccharides mitigate the adverse effects of iron fortification on the gut microbiome: a randomised controlled study in Kenyan infants. Gut 2017;66:1956-67.
4. De-Regil LM, Jefferds MED, Peña-Rosas JP. Point-of-use fortification of foods with micronutrient powders containing iron in children of preschool and school-age. Cochrane Database Syst Rev 2017;11:CD009666.
5. WHO. WHO guideline: use of multiple micronutrient powders for point-of-use fortification of foods consumed by infants and young children aged 6-23 months and children aged 2-12 years. 2016.
6. WHO. Nutritional anaemias: tools for effective prevention and control. 2017. Available from: https://www.who.int/publications/i/item/9789241513067. [Last accessed on 11 Dec 2024].
7. Puga AM, Samaniego-Vaesken ML, Montero-Bravo A, Ruperto M, Partearroyo T, Varela-Moreiras G. Iron supplementation at the crossroads of nutrition and gut microbiota: the state of the art. Nutrients 2022;14:1926.
8. Tang M, Frank DN, Hendricks AE, et al. Iron in micronutrient powder promotes an unfavorable gut microbiota in Kenyan infants. Nutrients 2017;9:776.
9. Jaeggi T, Kortman GAM, Moretti D, et al. Iron fortification adversely affects the gut microbiome, increases pathogen abundance and induces intestinal inflammation in Kenyan infants. Gut 2015;64:731-42.
10. Popovic A, Bourdon C, Wang PW, et al. Micronutrient supplements can promote disruptive protozoan and fungal communities in the developing infant gut. Nat Commun 2021;12:6729.
11. Mikulic N, Uyoga MA, Stoffel NU, et al. Prebiotics increase iron absorption and reduce the adverse effects of iron on the gut microbiome and inflammation: a randomized controlled trial using iron stable isotopes in Kenyan infants. Am J Clin Nutr 2024;119:456-69.
12. Rai D, Adelman AS, Zhuang W, Rai GP, Boettcher J, Lönnerdal B. Longitudinal changes in lactoferrin concentrations in human milk: a global systematic review. Crit Rev Food Sci Nutr 2014;54:1539-47.
13. Donker AE, van der Staaij H, Swinkels DW. The critical roles of iron during the journey from fetus to adolescent: developmental aspects of iron homeostasis. Blood Rev 2021;50:100866.
14. Oda H, Wakabayashi H, Yamauchi K, Abe F. Lactoferrin and bifidobacteria. Biometals 2014;27:915-22.
15. Gruden Š, Ulrih NP. Diverse mechanisms of antimicrobial activities of lactoferrins, lactoferricins, and other lactoferrin-derived peptides. Int J Mol Sci 2021;22:11264.
16. Siqueiros-Cendón T, Arévalo-Gallegos S, Iglesias-Figueroa BF, García-Montoya IA, Salazar-Martínez J, Rascón-Cruz Q. Immunomodulatory effects of lactoferrin. Acta Pharmacol Sin 2014;35:557-66.
17. Chen K, Chai L, Li H, et al. Effect of bovine lactoferrin from iron-fortified formulas on diarrhea and respiratory tract infections of weaned infants in a randomized controlled trial. Nutrition 2016;32:222-7.
18. Motoki N, Mizuki M, Tsukahara T, et al. Effects of lactoferrin-fortified formula on acute gastrointestinal symptoms in children aged 12-32 months: a randomized, double-blind, placebo-controlled trial. Front Pediatr 2020;8:233.
19. Mikulic N, Uyoga MA, Mwasi E, et al. Iron absorption is greater from apo-lactoferrin and is similar between holo-lactoferrin and ferrous sulfate: stable iron isotope studies in Kenyan infants. J Nutr 2020;150:3200-7.
20. ClinicalTrials.gov. Prebiotic GOS and lactoferrin with iron supplements. Available from: https://clinicaltrials.gov/study/NCT03866837. [Last accessed on 11 Dec 2024].
21. Vogt JA, Wolever TMS. Fecal acetate is inversely related to acetate absorption from the human rectum and distal colon. J Nutr 2003;133:3145-8.
22. Isenring J, Bircher L, Geirnaert A, Lacroix C. In vitro human gut microbiota fermentation models: opportunities, challenges, and pitfalls. Microbiome Res Rep 2023;2:2.
23. Zihler Berner A, Fuentes S, Dostal A, et al. Novel polyfermentor intestinal model (PolyFermS) for controlled ecological studies: validation and effect of pH. PLoS One 2013;8:e77772.
24. Doo EH, Chassard C, Schwab C, Lacroix C. Effect of dietary nucleosides and yeast extracts on composition and metabolic activity of infant gut microbiota in PolyFermS colonic fermentation models. FEMS Microbiol Ecol 2017;93:fix088.
25. Dostal A, Gagnon M, Chassard C, Zimmermann MB, O’Mahony L, Lacroix C. Salmonella adhesion, invasion and cellular immune responses are differentially affected by iron concentrations in a combined in vitro gut fermentation-cell model. PLoS One 2014;9:e93549.
26. Rachmühl C, Lacroix C, Cabrera PM, Geirnaert A. Long-term continuous cultivation of Kenyan infant fecal microbiota using the host adapted PolyFermS model. Sci Rep 2023;13:20563.
27. Cabrera P, Rachmühl C, Derrien M, Bourdet-Sicard R, Lacroix C, Geirnaert A. Comparative prebiotic potential of galacto- and fructo-oligosaccharides, native inulin, and acacia gum in Kenyan infant gut microbiota during iron supplementation. ISME Commun 2024;4:ycae033.
28. Poeker SA, Geirnaert A, Berchtold L, et al. Understanding the prebiotic potential of different dietary fibers using an in vitro continuous adult fermentation model (PolyFermS). Sci Rep 2018;8:4318.
29. Hernandez RJ, Gutowski D, Guire KE. Capacity of the colon in children. AJR Am J Roentgenol 1979;133:683-4.
30. Heinemann M, Strauchs C, Lütgehetmann M, et al. Polymicrobial enteric infections in African infants with diarrhoea-results from a longitudinal prospective case-control study. Clin Microbiol Infect 2021;27:1792-8.
31. Mikulic N, Uyoga MA, Paganini D, et al. Consumption of a single dose of prebiotic galacto-oligosaccharides does not enhance iron absorption from micronutrient powders in Kenyan infants: a stable iron isotope study. J Nutr 2021;151:1205-12.
32. Tondeur MC, Schauer CS, Christofides AL, et al. Determination of iron absorption from intrinsically labeled microencapsulated ferrous fumarate (sprinkles) in infants with different iron and hematologic status by using a dual-stable-isotope method. Am J Clin Nutr 2004;80:1436-44.
33. Walczyk T, Davidsson L, Zavaleta N, Hurrell RF. Stable isotope labels as a tool to determine the iron absorption by Peruvian school children from a breakfast meal. Fresenius J Anal Chem 1997;359:445-9.
34. Stoddard SF, Smith BJ, Hein R, Roller BR, Schmidt TM. rrnDB: improved tools for interpreting rRNA gene abundance in bacteria and archaea and a new foundation for future development. Nucleic Acids Res 2015;43:D593-8.
35. Callahan BJ, McMurdie PJ, Rosen MJ, Han AW, Johnson AJ, Holmes SP. DADA2: high-resolution sample inference from Illumina amplicon data. Nat Methods 2016;13:581-3.
37. Quast C, Pruesse E, Yilmaz P, et al. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res 2013;41:D590-6.
38. Vidal RM, Muhsen K, Tennant SM, et al. Colonization factors among enterotoxigenic Escherichia coli isolates from children with moderate-to-severe diarrhea and from matched controls in the Global Enteric Multicenter Study (GEMS). PLoS Negl Trop Dis 2019;13:e0007037.
39. McMurdie PJ, Holmes S. phyloseq: an R package for reproducible interactive analysis and graphics of microbiome census data. PLoS One 2013;8:e61217.
40. Oksanen AJ, Blanchet FG, Kindt R, et al. vegan: an R package for community ecologists. Available from: https://github.com/vegandevs/vegan. [Last accessed on 17 Dec 2024]
42. Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 2014;15:550.
43. Dostal A, Fehlbaum S, Chassard C, Zimmermann MB, Lacroix C. Low iron availability in continuous in vitro colonic fermentations induces strong dysbiosis of the child gut microbial consortium and a decrease in main metabolites. FEMS Microbiol Ecol 2013;83:161-75.
44. Celis AI, Relman DA, Huang KC. The impact of iron and heme availability on the healthy human gut microbiome in vivo and in vitro. Cell Chem Biol 2023;30:110-26.e3.
45. Fanaro S, Marten B, Bagna R, et al. Galacto-oligosaccharides are bifidogenic and safe at weaning: a double-blind randomized multicenter study. J Pediatr Gastroenterol Nutr 2009;48:82-8.
46. Scholtens PA, Alles MS, Bindels JG, van der Linde EG, Tolboom JJ, Knol J. Bifidogenic effects of solid weaning foods with added prebiotic oligosaccharides: a randomised controlled clinical trial. J Pediatr Gastroenterol Nutr 2006;42:553-9.
47. Souza DDS, Tahan S, Weber TK, Araujo-Filho HB, de Morais MB. Randomized, double-blind, placebo-controlled parallel clinical trial assessing the effect of fructooligosaccharides in infants with constipation. Nutrients 2018;10:1602.
48. Logtenberg MJ, Akkerman R, Hobé RG, et al. Structure-specific fermentation of galacto-oligosaccharides, isomalto-oligosaccharides and isomalto/malto-polysaccharides by infant fecal microbiota and impact on dendritic cell cytokine responses. Mol Nutr Food Res 2021;65:e2001077.
49. Le Blay G, Chassard C, Baltzer S, Lacroix C. Set up of a new in vitro model to study dietary fructans fermentation in formula-fed babies. Br J Nutr 2010;103:403-11.
50. Yao D, Wu M, Dong Y, et al. In vitro fermentation of fructooligosaccharide and galactooligosaccharide and their effects on gut microbiota and SCFAs in infants. J Funct Foods 2022;99:105329.
51. Martinez FA, Balciunas EM, Converti A, Cotter PD, de Souza Oliveira RP. Bacteriocin production by Bifidobacterium spp. a review. Biotechnol Adv 2013;31:482-8.
52. Vazquez-Gutierrez P, de Wouters T, Werder J, Chassard C, Lacroix C. High iron-sequestrating bifidobacteria inhibit enteropathogen growth and adhesion to intestinal epithelial cells in vitro. Front Microbiol 2016;7:1480.
53. McDonald JAK, Mullish BH, Pechlivanis A, et al. Inhibiting growth of clostridioides difficile by restoring valerate, produced by the intestinal microbiota. Gastroenterology 2018;155:1495-507.e15.
54. Hryckowian AJ, Van Treuren W, Smits SA, et al. Microbiota-accessible carbohydrates suppress Clostridium difficile infection in a murine model. Nat Microbiol 2018;3:662-9.
55. Kondepudi KK, Ambalam P, Nilsson I, Wadström T, Ljungh A. Prebiotic-non-digestible oligosaccharides preference of probiotic bifidobacteria and antimicrobial activity against Clostridium difficile. Anaerobe 2012;18:489-97.
56. Embleton N, Berrington J, Cummings S, Dorling J, Ewer A, Frau A, et al. Lactoferrin impact on gut microbiota in preterm infants with late-onset sepsis or necrotising enterocolitis: the MAGPIE mechanisms of action study. Effic Mech Eval 2021;8:1-88.
57. Sherman MP, Sherman J, Arcinue R, Niklas V. Randomized control trial of human recombinant lactoferrin: a substudy reveals effects on the fecal microbiome of very low birth weight infants. J Pediatr 2016;173:S37-42.
58. Chichlowski M, Bokulich N, Harris CL, et al. Effect of bovine milk fat globule membrane and lactoferrin in infant formula on gut microbiome and metabolome at 4 months of age. Curr Dev Nutr 2021;5:nzab027.
59. Young G, Berrington JE, Cummings S, et al. Mechanisms affecting the gut of preterm infants in enteral feeding trials: a nested cohort within a randomised controlled trial of lactoferrin. Arch Dis Child Fetal Neonatal Ed 2023;108:272-9.
60. Haiwen Z, Rui H, Bingxi Z, et al. Oral administration of bovine lactoferrin-derived lactoferricin (Lfcin) b could attenuate enterohemorrhagic Escherichia coli O157:H7 induced intestinal disease through improving intestinal barrier function and microbiota. J Agric Food Chem 2019;67:3932-45.
61. Teraguchi S, Shin K, Ozawa K, et al. Bacteriostatic effect of orally administered bovine lactoferrin on proliferation of Clostridium species in the gut of mice fed bovine milk. Appl Environ Microbiol 1995;61:501-6.
62. Rastogi N, Nagpal N, Alam H, et al. Preparation and antimicrobial action of three tryptic digested functional molecules of bovine lactoferrin. PLoS One 2014;9:e90011.
63. Lönnerdal B, Du X, Jiang R. Biological activities of commercial bovine lactoferrin sources. Biochem Cell Biol 2021;99:35-46.
64. Chilton CH, Crowther GS, Śpiewak K, et al. Potential of lactoferrin to prevent antibiotic-induced Clostridium difficile infection. J Antimicrob Chemother 2016;71:975-85.
65. Kortman GAM, Boleij A, Swinkels DW, Tjalsma H. Iron availability increases the pathogenic potential of Salmonella typhimurium and other enteric pathogens at the intestinal epithelial interface. PLoS One 2012;7:e29968.
66. Bhakat D, Mondal I, Mukhopadhyay AK, Chatterjee NS. Iron influences the expression of colonization factor CS6 of enterotoxigenic Escherichia coli. Microbiology 2021;167:001089.
67. Hinrichsen F, Hamm J, Westermann M, et al. Microbial regulation of hexokinase 2 links mitochondrial metabolism and cell death in colitis. Cell Metab 2021;33:2355-66.e8.
Cite This Article
How to Cite
Download Citation
Export Citation File:
Type of Import
Tips on Downloading Citation
Citation Manager File Format
Type of Import
Direct Import: When the Direct Import option is selected (the default state), a dialogue box will give you the option to Save or Open the downloaded citation data. Choosing Open will either launch your citation manager or give you a choice of applications with which to use the metadata. The Save option saves the file locally for later use.
Indirect Import: When the Indirect Import option is selected, the metadata is displayed and may be copied and pasted as needed.
About This Article
Copyright
Data & Comments
Data


























Comments
Comments must be written in English. Spam, offensive content, impersonation, and private information will not be permitted. If any comment is reported and identified as inappropriate content by OAE staff, the comment will be removed without notice. If you have any queries or need any help, please contact us at [email protected].