fig5

Figure 5. Correlations between maximum steady-state ATP hydrolysis rate and affinity in mouse embryo fibroblast membranes. (A): 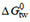 . The maximum steady-state ATP hydrolysis rate, V1, is expressed as a percentage of the basal steady-state ATP hydrolysis rate taken as 100%. Data were obtained from phosphate release measurements: (1) amitriptyline, (2) chlorpromazine, (3) cis-flupenthixol, (4) cyclosporine A, (5) daunorubicin, (6) dibucaine, (7) diltiazem, (8) glivec, (9) lidocaine, (10) OC144-093, (11) progesterone, (12) promazine, (13) reserpine, (14) trifluoperazine, (15) trifluopromazine, (16) PSC 833, (17) vinblastine (1-17, from Ref.[78]), (18) amlodipine, (19) nimodipine, (20) verapamil (18-20, from Ref. [93]), (21) sirolimus, (22) tacrolimus (21-22 from Simon Lang and A.S., unpublished results), (23) tariquidar (X. Li-Blatter and A.S., unpublished results), (24) etoposide (from Ref.[66]), (25) C12-maltoside, (26) C13-maltoside, (27) C12EO8, (28) Triton X-100, (29) Tween 80, (30) C10-TAC, (31) C12-TAC, (32) C14-TAC
. The maximum steady-state ATP hydrolysis rate, V1, is expressed as a percentage of the basal steady-state ATP hydrolysis rate taken as 100%. Data were obtained from phosphate release measurements: (1) amitriptyline, (2) chlorpromazine, (3) cis-flupenthixol, (4) cyclosporine A, (5) daunorubicin, (6) dibucaine, (7) diltiazem, (8) glivec, (9) lidocaine, (10) OC144-093, (11) progesterone, (12) promazine, (13) reserpine, (14) trifluoperazine, (15) trifluopromazine, (16) PSC 833, (17) vinblastine (1-17, from Ref.[78]), (18) amlodipine, (19) nimodipine, (20) verapamil (18-20, from Ref. [93]), (21) sirolimus, (22) tacrolimus (21-22 from Simon Lang and A.S., unpublished results), (23) tariquidar (X. Li-Blatter and A.S., unpublished results), (24) etoposide (from Ref.[66]), (25) C12-maltoside, (26) C13-maltoside, (27) C12EO8, (28) Triton X-100, (29) Tween 80, (30) C10-TAC, (31) C12-TAC, (32) C14-TAC 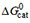 , and single HBA, as a function of the number of patterns per compound
, and single HBA, as a function of the number of patterns per compound 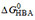 . Compound numbers and symbols as in (A), black open circles, uncharged compounds. The free energy per hydrogen bond (y-axis) decreases with the increasing number of hydrogen bonds in patterns per compound (x-axis). We extrapolated to one hydrogen bond in the case of electrically neutral compounds and compounds with low charge (red lines).
. Compound numbers and symbols as in (A), black open circles, uncharged compounds. The free energy per hydrogen bond (y-axis) decreases with the increasing number of hydrogen bonds in patterns per compound (x-axis). We extrapolated to one hydrogen bond in the case of electrically neutral compounds and compounds with low charge (red lines).












