Digital twins as a unifying framework for surgical data science: the enabling role of geometric scene understanding
Abstract
Surgical data science is devoted to enhancing the quality, safety, and efficacy of interventional healthcare. While the use of powerful machine learning algorithms is becoming the standard approach for surgical data science, the underlying end-to-end task models directly infer high-level concepts (e.g., surgical phase or skill) from low-level observations (e.g., endoscopic video). This end-to-end nature of contemporary approaches makes the models vulnerable to non-causal relationships in the data and requires the re-development of all components if new surgical data science tasks are to be solved. The digital twin (DT) paradigm, an approach to building and maintaining computational representations of real-world scenarios, offers a framework for separating low-level processing from high-level inference. In surgical data science, the DT paradigm would allow for the development of generalist surgical data science approaches on top of the universal DT representation, deferring DT model building to low-level computer vision algorithms. In this latter effort of DT model creation, geometric scene understanding plays a central role in building and updating the digital model. In this work, we visit existing geometric representations, geometric scene understanding tasks, and successful applications for building primitive DT frameworks. Although the development of advanced methods is still hindered in surgical data science by the lack of annotations, the complexity and limited observability of the scene, emerging works on synthetic data generation, sim-to-real generalization, and foundation models offer new directions for overcoming these challenges and advancing the DT paradigm.
Keywords
INTRODUCTION
Surgical data science is an emerging interdisciplinary research domain that has the potential to transform the future of surgery. Capitalizing on the pre- and intraoperative surgical data, the research efforts in surgical data science are dedicated to enhancing the quality, safety, and efficacy of interventional healthcare[1]. With the advent of powerful machine learning algorithms for surgical image and video analysis, surgical data science has witnessed a significant thrust, enabling solutions for problems that were once considered exceptionally difficult. These advances include improvements in low-level vision tasks, such as surgical instrument segmentation or “critical view of safety” classification[2], to high-level and downstream challenges, such as intraoperative guidance[3-7], intelligent assistance systems[8-10], surgical phase recognition[11-18], gesture classification[14,19-21], and skills analysis[20,22,23]. While end-to-end deep learning models have been the backbone of recent advancements in surgical data science, the high-level surgical analysis derived from these models raises concerns about reliability due to the lack of interpretability and explainability. Alternate to these end-to-end approaches, the emerging digital twin (DT) paradigm, a virtual equivalent of the real world, allows interpretable high-level surgical scene analysis on enriched digital data generated from low-level tasks.
End-to-end deep learning models have been the standard approach to surgical data science in both low- and high-level tasks. These models either focus on specific tasks or are used as foundational models, solving multiple downstream tasks. This somewhat straightforward approach, inspired by deep learning best practices, has historically excelled in task-specific performances due to deep learning’s powerful representation learning capabilities. However, we argue that this approach - despite its recent successes - is ripe for innovation[1,24,25]. End-to-end deep learning models exhibit strong tendencies to learn, exploit, or give in to non-causal relationships, or shortcuts, in the data[26-28]. Because it is impossible at worst or very difficult at best to distinguish low-level vision from high-level surgical data science components in end-to-end deep neural networks, it generally remains unclear how reliable these solutions are under various domain shifts and whether they associate the correct input signals with the resulting prediction[29]. These uncertainties and unreliability hinder further development in the surgical data science domain and the clinical translation of the current achievements[1]. While explainable machine learning, among other techniques, seeks to develop methods that may assert adequate model behavior[30], by and large, this limitation poses a challenge that we believe is not easily remedied with explanation-like constructs of similarly end-to-end deep learning origin.
The DT paradigm offers an alternative to task-specific end-to-end machine learning-based approaches for current surgical data science research. It provides a clear framework to separate low-level processing from high-level analysis. As a virtual equivalent of the real environment (surgical field in surgical data science), the DT models the real-world dynamics and properties using the data obtained from sensor-rich environments through low-level processing[7,31-33]. The resulting DT is ready for high-level complex analysis since all relevant quantities are known precisely and in a computationally accessible form. Unlike end-to-end deep learning paradigms that rely on data fitting, the DT paradigm employs data to construct a DT model. While the low-level processing algorithms that enable the DT are not immune to non-causal learning and environmental influences during the machine learning process, which might compromise robustness or performance, their impact is mostly confined to the accuracy of digital model construction and update. The resulting digital model in the DT paradigm can provide not only visual guidance like mixed reality but also, more importantly, a platform for more comprehensive surgical data science research like data generation, high-level surgical analysis (e.g., surgical phase recognition, and skill assessment), and autonomous agent training. DT’s uniform representation of causal factors, including geometric and physical attributes of the subjects and tools, surgery-related prior knowledge, and user input, along with their clear causal relation with the surgical task, should ensure superior generalizability and interpretability of surgical data science research.
The fundamental component of the DT paradigm is the building and updating of the digital model from real-world observations. In this process, geometric information processing plays a central role in the representation, visualization, and interaction of the digital model. Thus, the geometric scene understanding (i.e., perceiving geometric information from the target scene) is vital to enabling the realization of the DT and further DT-based research in surgical data science. In this work, we focus on reviewing the geometric representations, geometric scene understanding techniques, and their successful application for building primitive DT frameworks. The design of geometric representations needs to consider the trade-off among accuracy, representation ability, complexity, interactivity, and interpretability, as they correspond to the accuracy, applicability, efficiency, interactivity, and reliability of the DT framework. The extraction and encoding of different target representations and their status in the digital world lead to the establishment and development of various geometric scene understanding tasks, including segmentation, detection, depth estimation, 3D reconstruction, and pose estimation [Figure 1]. Although various sensors in a surgical setup, such as optical trackers, depth sensors, and robotics meters, are important for geometric understanding and DT instantiation in some procedures[7], we focus on reviewing methods based on visible light imaging, as it is the primary real-time observation source in most surgeries, especially in minimally invasive surgeries (MIS) due to their limited operational space. We further select methods that achieve superior benchmark performance for geometric scene understanding, which can be applied for accurate DT construction and updating. The integration of geometric scene understanding within the DT framework has led to successful applications, including simulator-empowered DT models and procedure-specific DT models.
Figure 1. The enabling role of geometric scene understanding for digital twin. All figures in the article are generated solely from the authors’ own resources, without any external references.
The paper is organized as follows: Section “GEOMETRIC REPRESENTATIONS” provides an overview of the existing digital representations for geometric understanding. Section “GEOMETRIC SCENE UNDERSTANDING TASKS” investigates existing datasets and various algorithms used to extract geometric understanding, assessing their effectiveness and limitations in terms of benchmark performance. Section “APPLICATIONS OF GEOMETRIC SCENE UNDERSTANDING EMPOWERED DIGITAL TWINS” explores the successful attempts to apply geometric scene understanding techniques in DT. Concluding the paper, Section “DISCUSSION” offers an in-depth discussion on the present landscape, challenges, and future directions in the field of geometric understanding within surgical data science.
GEOMETRIC REPRESENTATIONS
This section introduces various geometric representation categories and analyzes their advantages and disadvantages in relation to DT requirements. The taxonomy, along with some examples and summarized takeaways, is shown in Figure 2. We first present direct grid-based and point-based representations. Then, we discuss latent space representation generated via ruled encoding or neural encoding on previous representations or other modalities. Finally, we cover functional representations, which are generated through meticulous mathematical derivation or estimation based on observation.
Figure 2. An overview of geometric representations. PCA: Principal component analysis; SDF: signed distance function.
Grid-based representation
The grid-based representation divides the 2D/3D space into discrete cells and usually stores it as multidimensional arrays. Each cell holds values for various attributes, including density, color, semantic classes, and others. For example, the segmentation masks represent shapes of interest in rectangular space uniformly divided into a 2D grid of cells, where each cell is called a pixel. Some 3D shapes are also stored in uniformly divided 3D grids of cells. These cells are called voxels and usually hold the density/occupancy value. While these simplified representations enable efficient data queries for specific locations, they come with a trade-off between the error introduced by quantization and the computation and memory complexity[34]. The high computation and storage complexity hinders scaling up the accuracy and range of the representation. More importantly, there is an obvious gap between discrete representations and humans’ intuition of expressing geometry from a semantic level[34]. Humans perceive geometry in its entirety or connection between parts, whereas grid-based discrete representation represents geometry mostly by the spatial aggregation of the cells without explicit relations among them. Grid-based representations are suited well for tasks that focus on individual cell values/patterns in a group of cells. However, the lack of explicit relations among cells limits their geometric interpretability and their use in high-level processing tasks such as rigid/deformable surface representation and reconstruction.
Point-based representation
Instead of uniformly sampling the space, point-based representation samples key points represented in the Cartesian coordinate system.
Point cloud
A point cloud is a discrete set of data points in the 3D space[35]. It is the most basic point-based representation that can accurately represent the absolute position in an infinite space for each point. However, the sparsity of the points limits the accuracy of the representation at the object level. While it is easy to acquire from sensors, the lack of explicit relationships between points makes it slow to render. Similar to grid-based representation, the geometric interpretability of point clouds is limited due to the lack of relationships between points.
Boundaries
Boundaries are an alternate geometric representation method that introduces explicit relationships between points. An example in 3D space is the polygon mesh[36]. It is a collection of vertices, edges, and faces that define the shape of a polyhedral object. This type of representation is usually stored and processed as a graph structure. Boundary-based representations offer the flexibility to represent complex shapes and are widely used for 3D modeling. Compared to the point cloud, boundary-based representation offers faster rendering speed and more control over appearance[36]. A visual comparison is provided in Figure 3. However, it shares the same limitation as point clouds due to the sparsity of the points. Additionally, boundary-based representations have higher complexity and memory usage, as the potential connections increase quadratically as a function of the number of points. For high-level processing, the relation among points and explicit boundary representation provides basic geometric interpretability.
Latent space representation
Latent space representation is encoded from the original representation, such as point clouds, 2D images, or 3D volume, for dimension reduction or feature processing and aggregation. We divide the latent space representation into two subcategories based on the encoding methods.
Ruled encoding
Ruled encoding employs handcrafted procedures to encode the data to the latent space. One example is the principal component analysis (PCA)[37]. PCA linearly encodes the data to new coordinates according to the deviation of all samples. For geometric understanding, PCA can be used to extract the principal component as a model template for a specific object, e.g., face[38,39], and express the object as a linear combination of these templates. The latent representation reduces feature dimensionality and also removes noise[40]. However, the encoding is usually hard to interpret in geometric aspects and the projection and dimensional reduction usually come with information loss.
Neural encoding
Neural encoding extracts features via neural network architectures. The encoding is learned from specific data distributions with specific geometric signals or geometry-related tasks. Neural networks’ ability to extract higher-level features enhances the representation ability[41]. This makes neural encoding-based latent space representation work well when the downstream tasks are highly correlated to the pre-training proxy tasks and the application domain is similar to the training domain. However, when these conditions are not satisfied, the poor generalizability of neural networks[42] makes this type of representation less informative for downstream tasks. This limitation is also difficult to overcome due to the lack of geometric interpretability.
Functional representation
Functional representation captures geometric information using geometric constraints typically expressed as functions of the coordinates, specifying continuous sets of points/regions. We divide the functional representation into two subcategories based on the formation of the function.
Ruled functions
Ruled functions can be expressed in parametric ways or implicit ways. Parametric functions explicitly represent the point coordinates as a function of variables. Implicit functions take the point coordinates as input[43]. A signed distance function (SDF) f of spatial points x maps x to its orthogonal distance to the boundary of the represented object, where the sign is determined by whether or not x is in the interior of the object. The surface of the object is implicitly represented by the set {x | f(x) = 0}. Level sets represent geometric understanding by a function of coordinates. The value of the function is expressed as a level. The set of points that generate the same output value is a level set. A level set can be used to represent curves or surfaces. For primitive shapes like ellipses or rectangles, the ruled functional representation provides perfect accuracy, efficient storage, predictable manipulation, and quantized interpretability for some geometric properties. Thus, this representation form is commonly used in human-designed objects and simulations. However, for natural objects, although effort has been made to represent curves and surfaces using polynomials, as shown in the development of the Bezier/Berstein and B-spline theory[44], accurate parametric representations are still challenging to design.
Neural fields
Neural fields use the neural network’s universal approximation property[45] to approximate traditional functions to represent geometric information. For example, neural versions of level-set[46] and SDF[47] are proposed. Neural fields map the spatial points (and orientation) to specific attributes like colors (radiance field)[48] and signed orthogonal distances to the surface (SDF)[49]. Unlike latent feature representation, where the intermediate feature map extracted by the neural networks represents the geometric information, the Neural fields encode the geometric information into the networks’ parameters. Compared to the traditional parametric representation, the universal approximation ability[45] of neural networks enables the representation of complex geometric shapes and discontinuities learned from observations. The interpretability of the geometric information from the neural function depends on the function it approximates. For example, NeRF[48] lacks geometric interpretability as it models occupancy, whereas Neuralangelo[49] offers better geometric interpretability as it models SDF. However, since the network is a black box, geometric manipulation is not as direct as in ruled functional representations.
GEOMETRIC SCENE UNDERSTANDING TASKS
In this section, we visit and analyze some existing geometric scene understanding tasks and corresponding datasets and methods that are the building blocks of the DT framework. The illustration of tasks’ functionality and relative relation in the building and updating of DT is shown in Figure 4. We mainly introduce four large categories - segmentation and detection, depth estimation, 3D reconstruction, and pose estimation. Segmentation and detection in both static images and videos focus on identifying and isolating the target and generating decomposed semantic components from the entire scene. This procedure is the prerequisite for building and consistently updating the DT. While depth estimation extracts pixel-level depth information of the entire scene from static images, the generated depth map is limited in accuracy and representation ability due to its grid-based representation. Thus, it is not suitable as the only source for building and updating the digital twin and is often employed together with segmentation/detection methods and 3D reconstruction methods. Based on the identified targets, 3D reconstruction methods take multiple observations and extract more accurate and detailed geometric information to form 3D models for all components. Pose estimation tasks align the existing 3D models and their identification with the observations. While all techniques significantly contribute to building and updating the digital model, the building of the initial digital model mostly relies on segmentation, detection, and 3D reconstruction, adopting a “identify then reconstruct” principle. Once the digital model is initialized, the real-time updating of semantic components in the digital scene relies more on pose estimation. We first visit and summarize the availability of related data and materials in one subsection and then visit and analyze the techniques in the following subsections in the order of segmentation and detection, depth estimation, 3D reconstruction, and pose estimation.
Figure 4. An overview of geometric scene understanding tasks and corresponding impact in the building and updating of DT. All figures in the article are generated solely from the authors’ own resources, without any external references. DT: Digital twin.
Availability of data and materials
Segmentation and detection
Segmentation and detection, as fundamental tasks in surgical data science, receive an enormous amount of attention from the community. Challenges are being proposed with the corresponding dataset after the success of EndoVis challenges[50,51]. EndoVis[50,51] collected data from abdominal porcine procedures with the da Vinci surgical robot and manually annotated the surgical instruments and some tissue anatomy for segmentation. SAR-RARP50[52] and CATARACTS[53] challenge released in-vivo datasets for semantic segmentation in real procedures. SurgT[54] and STIR[55] provide annotations for tissue tracking. RobustMIS challenge[56] divided test data into three categories - same procedures as training, same surgery but different procedures, and different but similar surgery. With the three levels of similarity, the challenge aimed to assess algorithms’ robustness against domain shift. SegSTRONG-C[57] collected ex vivo data with manually added noise like smoke, blood, and low brightness to assess models’ robustness against non-adversarial corruptions unseen from the training data. Besides challenges, researchers also collect data to support algorithm development[58] and publications[59-61].
Although various datasets are available for segmentation and detection, due to the complexity of the segmentation and detection annotation, the scales (< 10k) of those datasets are not as large as the general vision data (MS COCO[62] 328k images, Object365[63], 600k images). Thus, SurgToolLoc[64] challenge provided tool presence annotation as weak labels in 24,695 video clips for machine learning models to be trained to detect and localize tools in video frames with bounding boxes. Lap. I2I Translation[65] attempted to generate a larger-scale dataset using an image-to-image translation approach from synthetic data. SegSTRONG-C[57] provided a foundation model-assisted[66] annotation protocol to expedite the annotation process. Despite the effort, the demand for a large-scale and uniform dataset exists desperately.
Depth estimation
Because of the difficulty in obtaining reliable ground truth, there are comparatively fewer surgical video datasets available for dense depth estimation. The EndoSLAM dataset consists of ex vivo and synthetic endoscopic video, with depth and camera pose information provided for each sample[67]. As a part of the Endoscopic Vision Challenge, the SCARED dataset includes porcine endoscopic video with ground truth obtained via structured light projection[68]. The Hamlyn Center dataset consists of in vivo laparoscopic and endoscopic videos, which are annotated in refs[69,70] with dense depth estimation using a pseudo-labeling strategy, resulting in the rectified Endo-Depth-and-Motion dataset (referred to as “Rectified Hamlyn”). Other smaller-scale datasets include the JHU Nasal Cavity dataset originally used for self-supervised monocular depth estimation[41], the Arthronet dataset[71], and two colonoscopy datasets with associated ground truth[72,73].
3D reconstruction
3D reconstruction relies on the correspondence among the observations of the same object/scene. Thus, unlike the datasets of other tasks where the image frames are always paired with ground truth annotations or weak annotations, any surgical video with adequate views of the target scene/objects can be used for 3D reconstruction. However, due to the limited observability in the surgical scenario, datasets with depth annotation[41,67,69,70,73] mentioned above or datasets containing stereo videos[57,74,75] are preferred for the 3D reconstruction research to overcome the ambiguity. Some datasets[67,73] also contain the ground-truth 3D model for the target anatomy for quantitative evaluation. Besides datasets already introduced in previous tasks, the JIGSAW[75] dataset, originally collected for surgical skill assessment, can be used for the reconstruction of surgical training scenarios.
Pose estimation
Pose estimation in laparoscopic surgery is important for accurate tool tracking and manipulation. SurgRIPE[76], part of the Endoscopic Vision Challenge 2023, addresses marker-less 6DoF pose estimation for surgical instruments under and without occlusion. The Laparoscopic Non-Robotic Dataset[77] focuses on simultaneous tool detection, segmentation, and geometric primitive extraction in laparoscopic non-robotic surgery. It includes tool presence, segmentation masks, and geometric primitives, supporting comprehensive tool detection and pose estimation tasks. Datasets capturing surgical tools in realistic conditions are essential for accurate pose estimation. The 3D Surgical Tools (3dStool) dataset[78] was constructed to include RGB images of surgical tools in action alongside their 3D poses. Four surgical tools were chosen: a scalpel, scissors, forceps, and an electric burr. The tools were recorded operating on a cadaveric knee to accurately mimic real-life conditions. The dVPose dataset[79] offers a realistic multi-modality dataset intended for the development and evaluation of real-time single-shot deep-learning-based 6D pose estimation algorithms on a head-mounted display. It includes comprehensive data for vision-based 6D pose estimation, featuring synchronized images from the RGB, depth, and grayscale cameras of the HoloLens 2 device, and captures the extra-corporeal portions of the instruments and endoscope of a da Vinci surgical robot.
Simulated and synthetic datasets can provide large-scale data and annotations through controlled environments. The Edinburgh Simulated Surgical Tools Dataset[80] includes RGBD images of five simulated surgical tools (two scalpels, two clamps, and one tweezer), featuring both synthetic and real images. Synthetic Surgical Needle 6DoF Pose Datasets[81] were generated with the AMBF simulator and assets from the Surgical Robotics Challenge[82]. These synthetic datasets focus on the 6DoF pose estimation of surgical needles, providing a controlled environment for developing and testing pose estimation algorithms. The POV-Surgery dataset[83] offers a large-scale, synthetic, egocentric collection focusing on pose estimation for hands with different surgical gloves and three orthopedic instruments: scalpel, friem, and diskplacer. It consists of high-resolution RGB-D video streams, activity annotations, accurate 3D and 2D annotations for hand-object poses, and 2D hand-object segmentation masks.
Pose estimation in surgical settings also extends to the use of X-ray imaging. i3PosNet[84] introduced three datasets: two synthetic Digitally Rendered Radiograph (DRR) datasets (one with a screw and the other with two surgical instruments) and a real X-ray dataset with manually labeled screws. These datasets facilitate the study and development of pose estimation algorithms using X-ray images in temporal bone surgery.
Table 1 summarizes the datasets mentioned in this section.
Summarization of datasets
| Category | Dataset |
| Segmentation and detection | EndoVis[50,51], Lap. I2I Translation[65], Sinus-Surgery-C/L[58], CholecSeg8k[59], CaDIS[53], RobustMIS[56], Kvasir-Instrument[60], AutoLaparo[61], SurgToolLoc[64], SAR-RARP50[52], SurT[54], STIR[55], SegSTRONG-C[57] |
| Depth estimation | EndoSLAM[67], SCARED[68], Rectified Hamlyn[70], JHU Nasal Cavity[41], Arthronet[71], AIST Colonoscopy[72], JHU Colonoscopy[73] |
| 3D reconstruction | JIGSAW[75], Lin et al.[74], Hamlyn[69], Rectified Hamlyn[70], EndoSLAM[67], JHU Nasal Cavity[41], JHU Colonoscopy[73] |
| Pose estimation | SurgRIPE[76], Laparoscopic Non-robotic Dataset[77], 3dStool[78], dVPose[79], Edinburgh Simulated Surgical Tools Dataset[80], Synthetic Surgical Needle 6DoF Pose Datasets[81], POV-Surgery[83], i3PosNet[84] |
Segmentation and detection
Segmentation and detection aim to identify the target objects from a static observation and represent them as space occupancy in pixel/voxel level labels or simplified bounding boxes for target instances, respectively. Segmentation and detection methods play a foundational role in performing geometric understanding for a complex scenario like surgery and provide basic geometric information by indicating which areas are occupied by objects. In this section, we focus on the segmentation and detection of videos and images. This section is divided into frame-wise and video-wise segmentation and detection, namely, (a) single frame segmentation and detection and (b) video object segmentation and tracking.
Single frame segmentation and detection
Starting from easier goals with the availability of more accurate and larger-scaled annotation, segmentation and detection in 2D space receive more attention. Due to different demands, the development of the detection and segmentation methods started from different perspectives. Traditional segmentation methods rely on low-level features such as edges and region colors, discriminating regions using thresholding or clustering techniques[85-87]. Traditional detection methods rely on human-crafted features and predict based on template matching[88-90]. With the emergence of deep learning algorithms, both detection and segmentation methods started using convolutional neural networks (CNNs) and adopted an end-to-end training regime.
In the general vision domain, segmentation was initially explored as a semantic segmentation task in the 2D space. As the target labels were generated from a fixed number of candidate classes, the output could be represented in the same format of multidimensional arrays as the input, making the encoder-decoder architecture-based FCN[91] a perfect fit for the task. Subsequent works[92-95] improved the performance by improving the feature aggregation techniques within the encoder-decoder architecture.
Object detection methods, unlike semantic segmentation, require the generation of rectangular bounding boxes for an arbitrary number of detected objects, in the format of a vector consisting of continuous center location and size that represents the spatial extent. The output representation that encompasses an arbitrary number of detected objects distinguishes instances from each other. As a consequence, the simple encoder-decoder architecture no longer meets the requirement. Instead, the detection pipeline based on region proposals represented by R-CNN[96-98] series and YOLO[99]/SSD[100] leads to the rapid emergence and development of two different paradigms of CNN-based object detection methods: two-stage paradigm[101-103] and one-stage paradigm[104-108].
The development paths for detection and segmentation converged with the need for instance-level segmentation. Instance segmentation requires a method to not only generate class labels for pixels but also distinguish instances from each other. Instance segmentation methods[109-112] that adopt a two-stage paradigm follow the “detect then segment” pipeline led by Mask R-CNN[109]. Single-stage paradigm methods[113-116] are free from generating bounding boxes first. The EndoVis instrument segmentation challenge and scene segmentation challenge[50,51] provide surgical-specific benchmarks and methods[117-122] targeting surgical video/images are proposed. These methods adopt architectures from the general computer vision domain, and some explore feature fusion mechanisms and data-inspired training techniques to better suit the training on surgical data. Meanwhile, due to the high-stakes nature of surgical procedures, the robustness of the segmentation and detection method is also an aspect that receives attention[2,56,123-125].
With the rise of vision transformers (ViT), ViT-based methods for segmentation and detection have received more attention. While numerous task-specific architectures have been introduced for semantic segmentation[126,127] and object detection[128-132], universal architectures/methods have also been explored[133,134]. Transformers architecture’s strong ability to deal with large-scale vision and language data and the effort of collecting immense amounts of data from industry lead to the rise of foundation models. CLIP[135] ignited the development of vision-language models[136-138] for segmentation and detection,
There are also methods designed for 3D segmentation or detection[148,149]. However, either their input representation is not visible light imaging like point cloud[150] or image including depth[151], or they require a 3D reconstruction from monocular[152] or multi-view 2D images[153] as a prerequisite for success. These are all beyond the scope of this paper or section.
Video object segmentation and tracking
With the introduction of strong computation resources and large-scale video-based annotation, the success of image-based segmentation and detection has led to the rise of video object segmentation and tracking. Video object segmentation and tracking aim to segment or track the objects with initial segmentation or detection results across the video sequence while maintaining their identities. The initial status can be given by an image-based segmentation or detection algorithm, or through manual annotation. Providing geometric understanding similar to that of instance-based segmentation and detection methods. Video object segmentation and tracking are vital for updating geometric understanding in a dynamic scene.
The development of video object segmentation algorithms mainly focuses on exploring the extraction and aggregation of spatial and temporal features for segmentation propagation and refinement. Temporal features are first explored with test-time online fine-tuning techniques[154,155] and recurrent approaches[156,157]. However, tracking under occlusions is challenging without spatial context. To address this, space-time memory[158-160] incorporates both spatial and temporal context with a running memory maintained for information aggregation. Exploiting the transformer architecture’s robust ability to deal with sequential data, such as videos, video ViT-based methods[161,162] have also showcased favorable results. Cutie[163] incorporates memory mechanism, transformer architecture, and feature fusion, achieving state-of-the-art results in video object segmentation.
The progression of video object tracking has witnessed the emergence of various task variants, such as video object detection[164-167], single object tracking[168-170], and multi-object tracking[171-173]. Traditional algorithms rely on sophisticated pipelines with handcrafted feature descriptors and optical flow for appearance modeling, smoothness assumptions-based motion modeling, object interaction, and occlusion handling, probabilistic inference, and deterministic optimization[174]. Applying neural networks simplifies the pipeline. One aspect of the effort focuses on modeling temporal feature flow or temporal feature aggregations[164-170] for better feature-level correspondence between frames. The other aspect focuses on improving object-level correspondence in multi-object scenarios[171-173].
Video object segmentation and tracking[175] in surgical videos mainly focus on moving objects like surgeons’ hands and surgical instruments. However, work focusing on tracking the tissues is still necessary when there is frequent camera movement. Taking advantage of a limited and sometimes fixed number of objects to track and similar appearance during the surgery, some traditional methods assume models of the targets[176-178], optimize with similarity functions or energy functions with respect to the observations and some low-level features, and apply registration or template matching for prediction. Some[179,180] rely on additional markers and other sensors for more accurate tracking.
Deep learning methods became the dominant approach once they were applied to surgical videos. Since less intra-class occlusion exists in the surgical scene, there is less demand for that sophisticated feature fusion mechanism in general vision. Most works continue using less data-consuming image-based segmentation and detection architectures[181-185] and maintain the correspondence at the result level. There are also works for end-to-end sequence models and spatial-temporal feature aggregation attempts[186,187].
Depth estimation
The goal of depth estimation is to associate with each pixel a value that reflects the distance from the camera to the object in that pixel, for a single timestep. This depth may be expressed in absolute units, such as meters, or in dimensionless units that capture the relative depth of objects in an image. The latter goal often arises for monocular depth estimation (MDE), which is depth estimation using a single image, due to the fundamental ambiguity between the scale of an object and its distance from the camera. Conventional approaches use shadows[188], edges, or structured light[189,190], to estimate relative depth. Although prior knowledge about the scale of specific objects in a scene enables absolute measurement, it was not until the advent of deep neural networks that object recognition became computationally tractable[191]. Stereo depth estimation (SDE), on the other hand, leverages the known geometry of multiple cameras to estimate depth in absolute units, and it has long been studied as a fundamental problem relevant to robotic navigation, manipulation, 3D modeling, surveying, and augmented reality applications[192,193]. Traditional approaches in this area used patch-based statistics[194] or edge detectors to identify sparse point correspondences between images, from which a dense disparity map can be interpolated[193,195]. Alternatively, dense correspondences can be established directly using local window-based techniques[196,197] or global optimization algorithms[198,199], which often make assumptions about surfaces being imaged to constrain the energy function[193]. However, the advent of deep learning revolutionized approaches to both monocular and stereo-depth estimation, proving particularly advantageous for the challenging domain of surgical images.
Within that domain, depth estimation is a valuable step toward geometric understanding in real time. As opposed to 3D reconstruction or structure-from-motion algorithms, as described below, depth estimation requires only a single frame of monocular or stereo video, meaning this geometric snapshot is just as reliable as post-hoc analysis that leverages future frames as well as past ones. Furthermore, it makes no assumptions about the geometric consistency of the anatomy throughout the surgery. In combination with detection and recognition algorithms, such as those above, depth estimation provides a semantically meaningful, 3D representation of the geometry of a surgical procedure, particularly minimally invasive procedures that rely on visual guidance. Traditionally, laparoscopic, endoscopic, and other visible spectrum image-guided procedures relied on monocular vision systems, which were sufficient when provided as raw guidance on a flat monitor[200,201]. The introduction of 3D monitors, which display stereo images using glasses to separate the left- and right-eye images, enabled stereo endoscopes and laparoscopes to be used clinically[202], proving to be invaluable to clinicians for navigating through natural orifices or narrow incisions[200,203]. Robot-assisted surgical robots like the da Vinci surgical system likewise use a 3D display in the surgeon console to provide a sense of depth. The increasing prevalence, therefore, of stereo camera systems in surgery has motivated the development of depth estimation algorithms for both modalities in this challenging domain.
Surgical video poses challenges for MDE in particular because it features deformable surfaces under variable lighting conditions, specular reflectances, and predominant motion along the non-ideal camera axis[204,205]. Deep neural networks (DNNs) offer a promising pathway for addressing these challenges by learning to regress dense depth maps consistently based on a priori knowledge of the target domain in the training set[206]. In this context, obtaining reliable ground truth is of the highest importance, and in general, this is obtained using non-monocular methods, while the DNN is restricted to analyzing the monocular image. Visentini-Scarzanella et al.[207] and Oda et al.[208], for example, obtained depth maps for colonoscopic video by rendering depth maps based on a reconstruction of the colon from CT. A more scalable approach uses the depth estimate from stereo laparoscopic[209] or arthroscopic[210] video, but this still requires stereo video data, which may not be available for procedures typically performed with monocular scopes. Using synthetic data rendered from photorealistic virtual models is a highly scalable method for generating images and depth maps, but DNNs must then overcome the sim-to-real gap[211-215]. Refs[204,216] explore the idea of self-supervised MDE by reconstructing the anatomical surface with structure-from-motion techniques but restricting the DNN to a single frame, later used widely[217,218]. Incorporating temporal consistency from recent frames can yield further improvements[219,220]. Datasets such as EndoSLAM[67] for endoscopy, SCARED[68] for laparoscopy, ref[71] for arthroscopy, and ref[221] for colonoscopy use these methods to make ground truth data more widely available for training[218,222-224]. For many of these methods, a straightforward DNN architecture with an encoder-decoder structure and straightforward loss function was used[204,211,212,216], although geometric constraints such as edge[208,225] or surface[223] consistency may be incorporated into the loss function. More drastic innovations explicitly confront the unique challenges of surgical videos, such as artificially removing smoke, which is frequently present due to cautery tools, using a combined GAN and U-Net architecture that simultaneously estimates depth[226].
As in detection and recognition tasks, large-scale foundation models have been developed with MDE in mind. The Depth-Anything model leverages both labeled (1.5M) and unlabeled (62M) images from real-world datasets to massively scale up the data available for training DNNs[227]. Rather than using structure-from-motion or other non-monocular methods to obtain ground truth for unlabeled data, Depth-Anything uses a previously obtained MDE teacher model to generate pseudo-labels for unlabeled images, preserving semantic features between the student and teacher models. Although trained using real-world images, Depth Anything’s zero-shot performance on endoscopic and laparoscopic video is nevertheless comparable to specialized models in terms of speed and performance[228]. It remains to be seen whether foundational models trained on real-world images will yield substantial benefits for surgical videos after fine-tuning or other transfer learning methods are explored.
For stereo depth estimation, DNNs have likewise shown vast improvements over conventional approaches. The ability of CNNs to extract salient local features has proved efficacious for establishing point correspondences based on image appearance, compared to handcrafted local or multi-scale window operators[229,230], leading to similar approaches on stereo laparoscopic video[231,232]. With regard to dense SDE, however, the ability of vision transformers[233] to train attention mechanisms on sequential information has proved especially apt for overcoming the challenges of generating globally consistent disparity maps, especially over smooth, deformable, or highly specular surfaces often encountered in surgical video[234-237]. When combined with object recognition and segmentation networks, as described above, they can be used to contend with significant occlusions such as those persistent in laparoscopic video from robotic-assisted minimally invasive surgery (RAMIS)[236].
3D reconstruction
Going a step beyond recognition, segmentation, and depth estimation, 3D reconstruction aims to generate explicit geometric information about a scene. In contrast to the depth estimation explained above, where the distance between the object and camera is represented as a distance value in a pixel, 3D reconstructed scenes are represented either using discrete representation (point cloud or mesh grid) or continuous representation (Neural Fields). In the visible light domain, 3D reconstruction refers to the intraoperative 3D reconstruction of surgical scenes, including anatomical tissues and surgical instruments. While it has been traditionally employed to reconstruct static tissues and organs, recently, novel techniques have been introduced for the 3D reconstruction of deformable tissues and to update preoperative 3D models based on intraoperative anatomical changes. Since most of the preoperative imaging modalities, such as CT and MRI, are 3D, inter-operative 3D reconstructive enables 3D-3D registration[41,210,238-241]. This makes real-time visible light imaging-based 3D reconstruction a key geometric understanding task that can aid surgical navigation, surgeon-centered augmented reality, and virtual reality[236].
3D reconstruction methods often use multiple images, acquired either altogether or at various times, to reconstruct a 3D model of the scene. Conventional reconstruction methods that estimate 3D structures from multiple 2D images include Structure from Motion (SfM)[242] and Simultaneous Localization and Mapping (SLAM)[243-247]. Similar to stereo depth estimation techniques, these methods fundamentally rely on motion parallax, the difference in object visualization from different image/camera viewpoints, for accurate estimation of the 3D structure of the object. One of the necessary tasks in estimating structure from motion is finding the correspondence between the different 2D images. Geometric information processing plays a key role in detecting and tracking features to establish correspondence. Such feature detection techniques include scale-invariant feature transform (SIFT) and Speeded-Up Robust Features (SURF). Alternatively, as visible light imaging-based surgical procedures are often equipped with stereo cameras, the use of depth estimation for the reconstruction of surgical scenes has also been reported[248]. Utilizing the camera pose information, the SLAM-based method allows the surgical scene reconstruction by fusing the depth information in the 3D space[244-246]. Although SfM and SLAM have shown promising performance in the natural computer vision domain, their application in the surgical domain has been limited, in part due to the paucity of features in the limited field of view. Additionally, these techniques assume the object to be static and rigid, which is not ideal for surgical scenes as the tissues/organs undergo deformations. The low-light imaging conditions, presence of bodily fluids, occlusions resulting from instrument movements, and specular reflections further affect the 3D reconstructions.
Discrete representation methods benefit from their sparsity properties, which improve efficiency in surface production. However, the same property also makes the representation method less robust in handling complex high-dimensional changes (non-topological deformations and color changes) - a general norm in surgical scenes – due to instrument-tissue interactions[249]. To address the deformations in tissue structures to an extent, sparse warp fields[250] have also been introduced in SuPer[251] and E-DSSR[236]. Novel techniques are also being explored for updating the preoperative CT models based on the soft tissue deformations and ablations observed through intraoperative endoscopic imaging[252]. Unlike discrete representation methods[248,249,253], emerging methods now employ continuous representations with the introduction of the Neural Radiance Field (NeRF) to reconstruct deformable tissues. Using the time-space input, the complex geometry and appearance are implicitly modeled to achieve high-quality 3D reconstruction[248,249]. EndoNeRF[248] employed two neural fields, where one is trained for tissue deformation and the other is trained for canonical density and appearance. It represents the deformable surgical scene as canonical neural radiance combined with a time-dependent neural displacement field. Advancing this further, EndoSurf[249] employs three neural fields to model the surgical dynamics, shape, and texture.
Pose estimation
Pose estimation aims to estimate the geometric relationship between an image and a prior model, which can take several forms[254]. These include rigid surface models, dense deformable surfaces, point-based skeletons, and robot kinematic models[254,255]. Many of the same techniques and variations employed in depth estimation and 3D reconstruction tasks also apply here, including feature-based matching with the perspective-n-point problem[255] and end-to-end learning-based approaches[256]. For point-based skeletons, such as the human body, pose estimation relied on handcrafted features[257] before the advent of deep neural networks[258]. In the context of surgical data science, pose estimation is highly relevant given the amount of a priori information going into any surgery, which can be leveraged to create 3D models. These include surgical tool models, robot models, and patient images. The 6DoF pose estimation of surgical tools, for example, in relation to patient anatomy, can enable algorithms that anticipate surgical errors and mitigate the risk of injuries[254]. By identifying tools’ proximity to critical structures, pose estimation technologies can ensure safer operations[259]. This is further advanced by precise 6DoF pose estimation of both instruments and tissue.
Deep neural networks have been shown to demonstrate promising outcomes for object pose estimation in RGB images[254,260-263]. Modern approaches often involve training models to regress 2D key points instead of directly estimating the object pose. These key points are then utilized to reconstruct the 6DoF object pose through the perspective-n-point (PnP) algorithm, with techniques showing robust performance, even in scenarios with occlusions[260].
Hand pose estimation also benefits from these technological advancements, with several methods proposed for deducing hand configurations from single-frame RGB images[264]. This capability is crucial for understanding the interactions between surgical tools and the operating environment, offering insights into the precise manipulation of instruments.
Beyond tool and hand pose estimation, human pose estimation can be applied for a broad spectrum of clinical applications, including surgical workflow analysis, radiation safety monitoring, and enhancing human-robot cooperation[265,266]. By leveraging videos from ceiling-mounted cameras, which capture both personnel and equipment in the operating room, human pose estimation can identify the finer activities within surgical phases, such as interactions between clinicians, staff, and medical equipment. The feasibility of estimating the poses of the individuals in an operating room, utilizing color images, depth images, or a combination of both, opens possibilities for real-time analysis of clinical environments[267].
APPLICATIONS OF GEOMETRIC SCENE UNDERSTANDING EMPOWERED DIGITAL TWINS
Geometric scene understanding plays a pivotal role in developing the DT framework by enabling the creation and real-time refinement of digital models based on real-world observations. Geometric information processing is crucial here for precise representation, visualization, and model interaction. Section “GEOMETRIC SCENE UNDERSTANDING TASKS” outlined the methods for processing this information, critical for navigating the complex geometry of surgical settings - identifying shapes, positions, and movements of anatomical features and tools. This section delves into the integration of geometric scene understanding within the DT framework, emphasizing its successful applications. It offers valuable insights that could be leveraged or specifically adapted to further the development of DT technologies in surgery.
Simulators serve as the essential infrastructure for creating, maintaining, and visualizing a DT of the physical world, crucially facilitating the collection and transmission of data back to the real world[268,269]. The Asynchronous Multi-Body Framework (AMBF) has demonstrated success in this regard through its applications in the surgical domain[270,271]. Building on this foundation, the fully immersive virtual reality for skull-base surgery (FIVRS) infrastructure, developed using AMBF, has been applied to DT frameworks and applications, demonstrating significant advancements in the field[7,272-275]. Twin-S, a DT framework designed for skull base surgery, leverages high-precision optical tracking and real-time simulation to model, track, and update the virtual counterparts of physical entities - such as the surgical drill, surgical phantom, tool-to-tissue interaction, and surgical camera - with high accuracy[7]. Additionally, this framework can be integrated with vision-based tracking algorithms[256], offering a potential alternative to optical trackers, thus enhancing its versatility and application scope. Contributing further to the domain, a collaborative robot framework has been developed to improve situational awareness in skull base surgery. This framework[274] introduces haptic assistive modes that utilize virtual fixtures based on generated signed distance fields (SDF) of critical anatomies from preoperative CT scans, thereby providing real-time haptic feedback. The effective communication between the real environment and the simulator is facilitated by adopting the Collaborative Robotics Toolkit (CRTK)[276] convention, which promotes modularity and seamless integration with other robotic systems. Additionally, an open-source toolset that integrates a physics-based constraint formulation framework, AMBF[270,271], with a state-of-the-art imaging platform application, 3D Slicer[277], has been developed[275]. This innovative toolset enables the creation of highly customizable interactive digital twins, incorporating the processing and visualization of medical imaging, robot kinematics, and scene dynamics for real-time robot control.
In addition to AMBF-empowered DT models, other DT models have also been proposed and explored for various surgical procedures. In liver surgery, a novel integration of thermal ablation with holographic augmented reality (AR) and DTs offers dynamic, real-time 3D navigation and motion prediction to improve accuracy and real-time performance[278]. Similarly, in the realm of cardiovascular interventions, the development of patient-specific artery models for coronary stenting simulations employs digital twins to personalize treatments. This approach uses finite element models derived from 3D reconstructions to validate in silico stenting procedures against actual clinical outcomes, underlining the move toward personalized care[279]. Orthopedic surgery benefits from applying DTs in evaluating the biomechanical effectiveness of stabilization methods for tibial plateau fractures generated from postoperative 3D X-ray images, aiding in optimizing surgical strategies and postoperative management[280]. The utilization of DT, AI, and machine learning to identify personalized motion axes for ankle surgery also marks a significant advancement, promising improvements in total ankle arthroplasty by ensuring the precise alignment of implants according to the specific anatomy of each patient[281]. Moreover, introducing a method to synchronize real and virtual manipulations in orthopedic surgeries through a dynamic DT enables surgeons to monitor and adjust the patient’s joint in real time with visual guidance. This technique not only ensures accurate alignments and adjustments during procedures but also significantly improves joint surgery outcomes.
The potential of DTs extends further when considering their role in enhancing higher-level downstream applications, ranging from surgical phase recognition and gesture classification to intraoperative guidance systems. Leveraging geometric understanding, DTs can interpret the broader context and flow of surgical operations, thereby increasing the precision and safety of interventions. Surgical phase recognition, for instance, utilizes insights from both direct video sources and interventional X-ray sequences to accurately identify the stages of a surgical procedure[11-18,282]. This facilitates a more structured and informed approach to surgery, enhancing the decision-making process and the efficacy of robotic assistants[14,19-21].
Furthermore, evaluating and enhancing surgical workflows and skills through these technologies can significantly advance surgeon training. By providing objective, quantifiable feedback on surgical techniques, DTs can support a comprehensive approach to assessing and improving surgical proficiency. This not only aids in training novices but also enhances performance evaluation across a spectrum of surgeons, from novices to experts, including those performing robot-assisted procedures[20,22,23,283-285]. The development of advanced cognitive surgical assistance technologies, based on the analysis of surgical workflows and skills, represents another opportunity. These technologies have the potential to enhance operational safety and foster semi-autonomous robotic systems that anticipate and adapt to the surgical team’s needs, thereby improving the collaborative efficiency of the surgical environment[1,3,286].
Intraoperative guidance technologies offer surgeons improved precision and real-time feedback[6]. Innovations such as mixed reality overlays and virtual fixtures could see their utility and efficacy greatly enhanced through integration with DTs. This synergy could further refine surgical accuracy and patient safety. Moreover, advancements like tool and needle guidance systems, alongside automated image acquisition[4,8], exemplify progress in geometric understanding and digital innovation. Integrating DTs with these surgical technologies holds the potential to improve standards for minimally invasive procedures and overall surgical quality.
Table 2 summarizes important methods for geometric scene understanding tasks and applications.
Summarization of methods
| Category | Methods |
| Segmentation and detection | EFFNet[117], Bamba et al.[118], Cerón et al.[119], Wang et al.[120], Zhang et al.[121], Yang et al.[122], CaRTS[2], TC-CaRTS[123], Colleoni et al.[124], daVinci GAN[125], AdptiveSAM[145], SurgicalSAM[146], Stenmark et al.[179], Cheng et al.[180], Zhao et al.[181,186], Robu et al.[182], Lee et al.[183], Seenivasan et al.[95], García-Peraza-Herrera et al.[184], Jo et al.[185], Alshirbaji et al.[187] |
| Depth estimation | Hannah[194], Marr and Poggio[192], Arnold[196], Okutomi and Kanade[197], Szeliski and Coughlan[198], Bouguet and Perona[188], Iddan and Yahav[189], Torralba and Oliva [191], Mueller-Richter et al.[201], Stoyanov et al.[195], Nayar et al.[190], Lo et al.[199], Sinha et al.[203], Liu et al.[206], Bogdanova et al.[202], Sinha et al.[200], Visentini-Scarzanella et al.[207], Mahmood et al.[211], Zhan et al.[209], Liu et al.[204], Guo et al.[212], Wong and Soatto[216], Chen et al.[213], Li et al.[205], Liu et al.[210], Schreiber et al.[214], Tong et al.[215], Widya et al.[217], Ozyoruk et al.[67], Allan et al.[68], Hwang et al.[219], Szeliski[193], Oda et al.[208], Shao et al.[218], Li et al.[220], Tukra and Giannarou[222], Ali and Pandey[71], Masuda et al.[221], Zhao et al.[223], Han et al.[224], Yang et al.[225], Zhang et al.[226] |
| 3D reconstruction | Dynamicfusion[250], Lin et al.[243], Song et al.[244], Zhou and Jagadeesan[245], Wdiya et al.[242], Li et al.[251], Wei et al.[247], EMDQ-SLAM[246], E-DSSR[236], EndoNeRF[248], EndoSurf[249], Nguyen et al.[253], Mangulabnan et al.[252] |
| Pose estimation | Hein et al.[254], Félix et al.[255], Tatoo[256], Allan et al.[259], Kadkhodamohammadi et al.[265], Padoy[266], Kadkhodamohammadi et al.[267] |
| Applications | FIVRS[272], Ishida et al.[273], Ishida et al.[274], Sahu et al.[275], Twin-S[7], Shi et al.[278], Poletti et al.[279], Aubert et al.[280], Hernigou et al.[281] |
DISCUSSION
The concept of DTs is rapidly gaining momentum in various surgical procedures, showcasing its transformative potential in shaping the future of surgery. The introduction of DT across various surgical procedures such as skull base surgery[7,272-275], liver surgery[278], cardiovascular intervention[279], and orthopedic surgery[280,281] demonstrate the broad effectiveness of DT technology in improving surgical precision and patient outcomes across different medical specialties. As stated in Section “APPLICATIONS OF GEOMETRIC SCENE UNDERSTANDING EMPOWERED DIGITAL TWINS”, the emergence of novel geometric scene understanding applications, such as phase recognition and gesture classification, could further empower DT models to interpret the broader context and flow of surgical operations. The potential ability to derive context-aware intelligence from a deep understanding of surgical dynamics further emphasizes DTs’ potential to advance robot-assisted surgeries and procedural planning. Geometric scene understanding-empowered DTs offer an alternate approach to the holistic understanding of the surgical scene through virtual models and have the potential to subsequently enhance the surgical process, from planning and execution to training and postoperative analysis, driving the digital revolution in surgery.
Geometric scene understanding forms the backbone of DTs that incorporate and process diverse enriched data from different stages of surgery. The evolution of geometric information processing has transitioned from simple low-level feature processing to neural network-based methods, introducing innovative geometric representations like neural fields. Various geometric scene understanding tasks have also been established, with corresponding methods achieving significant performance improvements. However, challenges persist in the representation of geometric information, the development of geometric scene understanding within the surgical domain, and its application to the DT paradigm.
In geometric representations, there is no single form that can meet all the requirements of DT in terms of accuracy, applicability, efficiency, interactivity, and reliability. Grid-based methods compromise the accuracy and processing efficiency for convenient structure and representation ability. It also lacks interactivity and reliability, as the geometric information is mainly represented by the aggregation of adjacent pixels. Point-based representations provide accurate positions for each point. It also allows the establishment of explicit connections between points to form boundaries, improving rendering efficiency and providing more geometric constraints for interaction and interpretation. Thus, polygon mesh representation has been widely used in 3D surface reconstruction and digital modeling. However, it is computationally less efficient, and the sparsity of the points still limits the accuracy. Latent space representation, while offering the possibility of dimensional reduction (ruled encoding) for efficiency and high-level semantic incorporation (neural encoding), lacks accuracy due to information loss and reliability due to limited interpretability and generalizability. Functional representations offer a new approach to representing geometric information through mathematical constraints or mappings. Ruled functions have the advantages of processing efficiency, easy interactivity, and quantized interpretability. However, it lacks the ability to represent complex surfaces. On the other hand, the neural fields[48,49], leveraging on neural networks’ universal approximation ability, demonstrate remarkable representation capabilities. This enables the efficient continuous 3D representation of complex and dynamic surgical scenes, which makes it a popular topic. However, the use of black-box networks sacrifices interactivity and interpretability. While no one geometric representation is optimal for all cases, the current advances require the user to choose data presentation based on the trade-off and the task-specific priority. For tasks that require robust performance in all aspects, future work could explore novel data representation that fuses sparse representations and neural fields, to achieve better surface representation with lesser computation load.
Geometric scene understanding has made immense strides in the general computer vision domain. However, its progress in the surgical domain is hindered by multiple factors. Firstly, limited data availability and annotations have become a major roadblock in adapting advanced but data-consuming architectures like ViT[233] from the computer vision domain. This significantly impacts the accuracy and reliability of segmentation and detection, which are prerequisites for the success of DT. Although self-supervised techniques of using stereo matching exist that might exempt depth estimation from lack of annotations, the stability[287] of training needs careful attention. While efforts have been made to bridge the gap in the scale of data between computer vision and the surgical domain through synthetic data generation and sim-to-real generalization techniques[288,289], this direction also poses challenges due to the lack of interpretability for neural networks. Secondly, the complexity of the surgical scene, including non-static organs and deformable tissues, poses another major challenge when updating the DT models relies solely on pose estimation, with the assumption that 3D models are rigid. Although dynamic 3D reconstruction methods exist[248,249], they cannot currently be processed in real time for updating geometric information and require auxiliary constraints like stereo matching for plausible output. The lack of observability due to limited operational space is also a major challenge that leads to a paucity of features for geometric scene understanding. Incorporating other modalities like robot kinematics[2] and temporal constraints[123] can be a complement under this situation. The emergence of foundation models also offers an alternate approach to harness the power of foundation models trained on enormous natural images to address the aforementioned challenges in the surgical domain[146,145]. However, the domain gap may hinder the optimal extraction of precise features[147] and may require further work to extend them in the surgical domain.
CONCLUSION
Surgical data science, benefiting from the advent of end-to-end deep learning architectures, is also hindered by their lack of reliability and interoperability. The DT paradigm is envisioned to advance the surgical data science domain further, introducing new avenues of research in surgical planning, execution, training, and postoperative analysis by providing a universal digital representation that enables robust and interpretable surgical data science research. Geometric scene understanding is the core building block of DT and plays a pivotal role in building and updating digital models. In this review, we find that the existing geometric representation and well-established tasks provide fundamental materials and tools to implement the DT framework and have led to the emergence of successful applications. However, challenges remain in employing more advanced but data-consuming methods especially in segmentation, detection, and monocular depth estimation tasks in the surgical domain due to a lack of annotations and a gap in the scale of the data. The complexity of the surgical scene due to the large portion of dynamic and deformable tissues, and the lack of observability due to limited operational space are also common factors that hinder the development of geometric scene understanding tasks, especially for the 3D reconstruction that demands multi-view observations. To address these challenges, numerous approaches, including synthetic image generation, sim-to-real generalization, auxiliary data incorporation, and foundational model adaptation, are being explored. Among all of these methods, the auxiliary data incorporation and foundation models present the most promising improvement. Since the auxiliary data is not always available and the exploration of the foundation models in surgical data science is still preliminary, it is expected to see more advancement in this direction that improves the geometric scene understanding performance and further promotes DT research. Developing an accurate, efficient, interactive, and reliable DT requires robust and efficient holistic geometric representation and combinations of effective geometric scene understanding, to build and update digital model pipelines in real time.
DECLARATIONS
Authors’ contributions
Initial writing of the majority part and coordination of the collaboration among authors: Ding H
Initial writing of the 3D reconstruction, integration, and revision of the paper: Seenivasan L
Initial writing of the depth estimation and pose estimation part and revision of the paper: Killeen BD
Initial writing of the pose estimation and application part: Cho SM
The main idea of the paper, overall structure, and revision of the paper: Unberath M
Availability of data and materials
See Section “Availability of data and materials” in the main text.
Financial support and sponsorship
This research is in part supported by (1) the collaborative research agreement with the Multi-Scale Medical Robotics Center at The Chinese University of Hong Kong; (2) the Link Foundation Fellowship for Modeling, Training, and Simulation; and (3) NIH R01EB030511 and Johns Hopkins University Internal Funds. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Conflict of interest
All authors declared that there are no conflicts of interest.
Ethical approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Copyright
© The Author(s) 2024.
REFERENCES
1. Maier-Hein L, Eisenmann M, Sarikaya D, et al. Surgical data science - from concepts toward clinical translation. Med Image Anal. 2022;76:102306.
2. Ding H, Zhang J, Kazanzides P, Wu JY, Unberath M. CaRTS: causality-driven robot tool segmentation from vision and kinematics data. In: Wang L, Dou Q, Fletcher PT, Speidel S, Li S, editors. Medical Image Computing and Computer Assisted Intervention - MICCAI 2022. Cham: Springer; 2022. pp. 387-98.
3. Kenngott HG, Wagner M, Preukschas AA, Müller-Stich BP. [Intelligent operating room suite: from passive medical devices to the self-thinking cognitive surgical assistant]. Chirurg. 2016;87:1033-8.
4. Killeen BD, Gao C, Oguine KJ, et al. An autonomous X-ray image acquisition and interpretation system for assisting percutaneous pelvic fracture fixation. Int J Comput Assist Radiol Surg. 2023;18:1201-8.
5. Gao C, Killeen BD, Hu Y, et al. Synthetic data accelerates the development of generalizable learning-based algorithms for X-ray image analysis. Nat Mach Intell. 2023;5:294-308.
6. Madani A, Namazi B, Altieri MS, et al. Artificial intelligence for intraoperative guidance: using semantic segmentation to identify surgical anatomy during laparoscopic cholecystectomy. Ann Surg. 2022;276:363-9.
7. Shu H, Liang R, Li Z, et al. Twin-S: a digital twin for skull base surgery. Int J Comput Assist Radiol Surg. 2023;18:1077-84.
8. Killeen BD, Winter J, Gu W, et al. Mixed reality interfaces for achieving desired views with robotic X-ray systems. Comput Methods Biomech Biomed Eng Imaging Vis. 2023;11:1130-5.
9. Killeen BD, Chaudhary S, Osgood G, Unberath M. Take a shot! Natural language control of intelligent robotic X-ray systems in surgery. Int J Comput Assist Radiol Surg. 2024;19:1165-73.
10. Kausch L, Thomas S, Kunze H, et al. C-arm positioning for standard projections during spinal implant placement. Med Image Anal. 2022;81:102557.
11. Killeen BD, Zhang H, Mangulabnan J, et al. Pelphix: surgical phase recognition from X-ray images in percutaneous pelvic fixation. In: Greenspan H, Madabhushi A, Mousavi P, Salcudean S, Duncan J, Syeda-mahmood T, Taylor R, editors. Medical Image Computing and Computer Assisted Intervention - MICCAI 2023. Cham: Springer; 2023. pp. 133-43.
12. Garrow CR, Kowalewski KF, Li L, et al. Machine learning for surgical phase recognition: a systematic review. Ann Surg. 2021;273:684-93.
13. Weede O, Dittrich F, Worn H, et al. Workflow analysis and surgical phase recognition in minimally invasive surgery. In: 2012 IEEE International Conference on Robotics and Biomimetics (ROBIO); 2012 Dec 11-14; Guangzhou, China. IEEE; 2012. pp. 1080-74.
14. Kiyasseh D, Ma R, Haque TF, et al. A vision transformer for decoding surgeon activity from surgical videos. Nat Biomed Eng. 2023;7:780-96.
15. Ban Y, Eckhoff JA, Ward TM, et al. Concept graph neural networks for surgical video understanding. IEEE Trans Med Imaging. 2024;43:264-74.
16. Czempiel T, Paschali M, Keicher M, et al. TeCNO: surgical phase recognition with multi-stage temporal convolutional networks. In: Martel AL, Abolmaesumi P, Stoyanov D, Mateus D, Zuluaga MA, Zhou SK, Racoceanu D, Joskowicz L, editors. Medical Image Computing and Computer Assisted Intervention - MICCAI 2020. Cham: Springer; 2020. pp. 343-52.
17. Guédon ACP, Meij SEP, Osman KNMMH, et al. Deep learning for surgical phase recognition using endoscopic videos. Surg Endosc. 2021;35:6150-7.
18. Murali A, Alapatt D, Mascagni P, et al. Encoding surgical videos as latent spatiotemporal graphs for object and anatomy-driven reasoning. In: Greenspan H, et al., editors. Medical Image Computing and Computer Assisted Intervention - MICCAI 2023. Cham: Springer; 2023. pp. 647-57.
19. Zhang D, Wang R, Lo B. Surgical gesture recognition based on bidirectional multi-layer independently RNN with explainable spatial feature extraction. In: 2021 IEEE International Conference on Robotics and Automation (ICRA); 2021 May 30 - Jun 5; Xi’an, China. IEEE; 2021. pp. 1350-6.
20. DiPietro R, Ahmidi N, Malpani A, et al. Segmenting and classifying activities in robot-assisted surgery with recurrent neural networks. Int J Comput Assist Radiol Surg. 2019;14:2005-20.
21. Dipietro R, Hager GD. Automated surgical activity recognition with one labeled sequence. In: Shen D, et al., editors. Medical Image Computing and Computer Assisted Intervention - MICCAI 2019. Cham: Springer; 2019. pp. 458-66.
22. Reiley CE, Lin HC, Yuh DD, Hager GD. Review of methods for objective surgical skill evaluation. Surg Endosc. 2011;25:356-66.
23. Lam K, Chen J, Wang Z, et al. Machine learning for technical skill assessment in surgery: a systematic review. NPJ Digit Med. 2022;5:24.
24. Alapatt D, Murali A, Srivastav V, Mascagni P, Consortium A, Padoy N. Jumpstarting surgical computer vision. arXiv. [Preprint.] Dec 10, 2023 [accessed 2024 Jul 2]. Available from: https://arxiv.org/abs/2312.05968.
25. Ramesh S, Srivastav V, Alapatt D, et al. Dissecting self-supervised learning methods for surgical computer vision. Med Image Anal. 2023;88:102844.
26. Geirhos R, Rubisch P, Michaelis C, Bethge M, Wichmann FA, Brendel W. Imagenet-trained cnns are biased towards texture; increasing shape bias improves accuracy and robustness. arXiv. [Preprint.] Nov 29, 2018 [accessed 2024 Jul 2]. Available from: https://arxiv.org/abs/1811.12231.
27. Glocker B, Jones C, Roschewitz M, Winzeck S. Risk of bias in chest radiography deep learning foundation models. Radiol Artif Intell. 2023;5:e230060.
28. Geirhos R, Jacobsen J, Michaelis C, et al. Shortcut learning in deep neural networks. Nat Mach Intell. 2020;2:665-73.
29. Wen C, Qian J, Lin J, Teng J, Jayaraman D, Gao Y. Fighting fire with fire: avoiding dnn shortcuts through priming. Available from: https://proceedings.mlr.press/v162/wen22d.html. [Last accessed on 2 Jul 2024]
30. Olah C, Satyanarayan A, Johnson I, et al. The building blocks of interpretability. Distill. 2018;3:e10.
32. Bjelland Ø, Rasheed B, Schaathun HG, et al. Toward a digital twin for arthroscopic knee surgery: a systematic review. IEEE Access. 2022;10:45029-52.
33. Erol T, Mendi AF, Doğan D. The digital twin revolution in healthcare. In: 2020 4th International Symposium on Multidisciplinary Studies and Innovative Technologies (ISMSIT); 2020 Oct 22-24; Istanbul, Turkey. IEEE; 2020. pp. 1-7.
34. Representations of geometry for computer graphics. Available from: https://graphics.stanford.edu/courses/cs233-24-winter-v1/ReferencedPapers/60082881-Presentations-of-Geometry-for-Computer-Graphics.pdf. [Last accessed on 2 Jul 2024].
35. Levoy M, Whitted T. The use of points as a display primitive. 2000. Available from: https://api.semanticscholar.org/CorpusID:12672240. [Last accessed on 2 Jul 2024].
36. Botsch M, Kobbelt L, Pauly M, Alliez P, Levy B. Polygon mesh processing. A K Peters/CRC Press; 2010. Available from: http://www.crcpress.com/product/isbn/9781568814261. [Last accessed on 2 Jul 2024]
37. Jolliffe IT, Cadima J. Principal component analysis: a review and recent developments. Philos Trans A Math Phys Eng Sci. 2016;374:20150202.
38. Blanz V, Vetter T. A morphable model for the synthesis of 3D faces. In: Whitton MC, editor. Seminal graphics papers: pushing the boundaries. New York: ACM; 2023. pp. 157-64.
39. Edwards GJ, Taylor CJ, Cootes TF. Interpreting face images using active appearance models. In: Proceedings Third IEEE International Conference on Automatic Face and Gesture Recognition; 1998 Apr 14-16; Nara, Japan. IEEE; 1998. pp. 300-5.
40. Karamizadeh S, Abdullah SM, Manaf AA, Zamani M, Hooman A. An overview of principal component analysis. J Signal Inf Process. 2013;4:173-5.
41. Liu X, Killeen BD, Sinha A, et al. Neighborhood normalization for robust geometric feature learning. In: 2021 IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR); 2021 Jun 20-25; Nashville, TN, USA. IEEE; 2021. pp. 13049-58.
42. Drenkow N, Sani N, Shpitser I, Unberath M. A systematic review of robustness in deep learning for computer vision: mind the gap? arXiv. [Preprint.] Dec 1, 2021 [accessed 2024 Jul 2]. Available from: https://arxiv.org/abs/2112.00639.
45. Hornik K, Stinchcombe M, White H. Multilayer feedforward networks are universal approximators. Neural Netw. 1989;2:359-66.
46. Michalkiewicz M, Pontes JK, Jack D, Baktashmotlagh M, Eriksson A. Deep level sets: implicit surface representations for 3d shape inference. arXiv. [Preprint.] Jan 21, 2019 [accessed 2024 Jul 2]. Available from: https://arxiv.org/abs/1901.06802.
47. Park JJ, Florence P, Straub J, Newcombe R, Lovegrove S. Deepsdf: learning continuous signed distance functions for shape representation. arXiv. [Preprint.] Jan 16, 2019 [accessed 2024 Jul 2]. Available from: https://arxiv.org/abs/1901.05103.
48. Mildenhall B, Srinivasan PP, Tancik M, Barron JT, Ramamoorthi R, Ng R. NeRF: representing scenes as neural radiance fields for view synthesis. Commun ACM. 2022;65:99-106.
49. Li Z, Müller T, Evans A, et al. Neuralangelo: high-fidelity neural surface reconstruction. In: 2023 IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR); 2023 Jun 17-24; Vancouver, BC, Canada. IEEE; 2023. pp. 8456-65.
50. Allan M, Shvets A, Kurmann T, et al. 2017 robotic instrument segmentation challenge. arXiv. [Preprint.] Feb 21, 2019 [accessed 2024 Jul 2]. Available from: https://arxiv.org/abs/1902.06426.
51. Allan M, Kondo S, Bodenstedt S, et al. 2018 robotic scene segmentation challenge. arXiv. [Preprint.] Aug 3, 2020 [accessed 2024 Jul 2]. Available from: https://arxiv.org/abs/2001.11190.
52. Psychogyios D, Colleoni E, Van Amsterdam B, et al. Sar-rarp50: segmentation of surgical instrumentation and action recognition on robot-assisted radical prostatectomy challenge. arXiv. [Preprint.] Jan 23, 2024 [accessed 2024 Jul 2]. Available from: https://arxiv.org/abs/2401.00496.
53. Grammatikopoulou M, Flouty E, Kadkhodamohammadi A, et al. CaDIS: Cataract dataset for surgical RGB-image segmentation. Med Image Anal. 2021;71:102053.
54. Cartucho J, Weld A, Tukra S, et al. SurgT challenge: benchmark of soft-tissue trackers for robotic surgery. Med Image Anal. 2024;91:102985.
55. Schmidt A, Mohareri O, DiMaio S, Salcudean SE. Surgical tattoos in infrared: a dataset for quantifying tissue tracking and mapping. arXiv. [Preprint.] Feb 29, 2024 [accessed 2024 Jul 2]. Available from: https://arxiv.org/abs/2309.16782.
56. Roß T, Reinke A, Full PM, et al. Comparative validation of multi-instance instrument segmentation in endoscopy: Results of the ROBUST-MIS 2019 challenge. Med Image Anal. 2021;70:101920.
57. Segstrong-C: segmenting surgical tools robustly on non-adversarial generated corruptions - an EndoVis’24 challenge. [Preprint.] Jul 16, 2024 [accessed 2024 Jul 18]. Available from: https://arxiv.org/abs/2407.11906.
58. Qin F, Lin S, Li Y, Bly RA, Moe KS, Hannaford B. Towards better surgical instrument segmentation in endoscopic vision: multi-angle feature aggregation and contour supervision. IEEE Robot Autom Lett. 2020;5:6639-46.
59. Hong WY, Kao CL, Kuo YH, Wang JR, Chang WL, Shih CS. Cholecseg8k: a semantic segmentation dataset for laparoscopic cholecystectomy based on cholec80. arXiv. [Preprint.] Dec 23, 2020 [accessed 2024 Jul 2]. Available from: https://arxiv.org/abs/2012.12453.
60. Jha D, Ali S, Emanuelsen K, et al. Kvasir-instrument: diagnostic and therapeutic tool segmentation dataset in gastrointestinal endoscopy. In: Lokoč J, et al., editors. MultiMedia Modeling. Cham: Springer; 2021. pp. 218-29.
61. Wang Z, Lu B, Long Y, et al. AutoLaparo: a new dataset of integrated multi-tasks for image-guided surgical automation in laparoscopic hysterectomy. In: Wang L, Dou Q, Fletcher PT, Speidel S, Li S, editors. Medical Image Computing and Computer Assisted Intervention - MICCAI 2022. Cham: Springer; 2022. pp. 486-96.
62. Lin TY, Maire M, Belongie S, et al. Microsoft COCO: common objects in context. In: Fleet D, Pajdla T, Schiele B, Tuytelaars T, editors. Computer Vision - ECCV 2014. Cham: Springer; 2014. pp. 740-55.
63. Shao S, Li Z, Zhang T, et al. Objects365: a large-scale, high-quality dataset for object detection. In: 2019 IEEE/CVF International Conference on Computer Vision (ICCV); 2019 Oct 27 - Nov 2; Seoul, Korea (South). IEEE; 2019. pp. 8429-38.
64. Zia A, Bhattacharyya K, Liu X, et al. Surgical tool classification and localization: results and methods from the miccai 2022 surgtoolloc challenge. arXiv. [Preprint.] May 31, 2023 [accessed 2024 Jul 2]. Available from: https://arxiv.org/abs/2305.07152.
65. Pfeiffer M, Funke I, Robu MR, et al. Generating large labeled data sets for laparoscopic image processing tasks using unpaired image-to-image translation. In: Shen D, et al., editors. Medical Image Computing and Computer Assisted Intervention - MICCAI 2019. Cham: Springer; 2019. pp. 119-27.
66. Kirillov A, Mintun E, Ravi N, et al. Segment anything. arXiv. [Preprint.] Apr 5, 2023 [accessed 2024 Jul 2]. Available from: https://arxiv.org/abs/2304.02643.
67. Ozyoruk KB, Gokceler GI, Bobrow TL, et al. EndoSLAM dataset and an unsupervised monocular visual odometry and depth estimation approach for endoscopic videos. Med Image Anal. 2021;71:102058.
68. Allan M, Mcleod J, Wang C, et al. Stereo correspondence and reconstruction of endoscopic data challenge. arXiv. [Preprint.] Jan 28, 2021 [accessed 2024 Jul 2]. Available from: https://arxiv.org/abs/2101.01133.
69. Hamlyn centre laparoscopic/endoscopic video datasets. Available from: https://hamlyn.doc.ic.ac.uk/vision/. [Last accessed on 2 Jul 2024].
70. Recasens D, Lamarca J, Fácil JM, Montiel JMM, Civera J. Endo-depth-and-motion: reconstruction and tracking in endoscopic videos using depth networks and photometric constraints. IEEE Robot Autom Lett. 2021;6:7225-32.
71. Ali S, Pandey AK. ArthroNet: a monocular depth estimation technique with 3D segmented maps for knee arthroscopy. Intell Med. 2023;3:129-38.
72. Masuda K, Shimizu T, Nakazawa T, Edamoto Y. Registration between 2D and 3D ultrasound images to track liver blood vessel movement. Curr Med Imaging. 2023;19:1133-43.
73. Bobrow TL, Golhar M, Vijayan R, Akshintala VS, Garcia JR, Durr NJ. Colonoscopy 3D video dataset with paired depth from 2D-3D registration. Med Image Anal. 2023;90:102956.
74. Lin B, Sun Y, Sanchez JE, Qian X. Efficient vessel feature detection for endoscopic image analysis. IEEE Trans Biomed Eng. 2015;62:1141-50.
75. JIGSAWS: the JHU-ISI gesture and skill assessment working set: JHU-ISI Gesture and skill assessment working set (JIGSAWS). Available from: https://cirl.lcsr.jhu.edu/research/hmm/datasets/jigsaws_release/. [Last accessed on 2 Jul 2024].
76. Hein J, Cavalcanti N, Suter D, et al. Next-generation surgical navigation: marker-less multi-view 6dof pose estimation of surgical instruments. arXiv. [Preprint.] Dec 22, 2023 [accessed 2024 Jul 2]. Available from: https://arxiv.org/abs/2305.03535.
77. Hasan MK, Calvet L, Rabbani N, Bartoli A. Detection, segmentation, and 3D pose estimation of surgical tools using convolutional neural networks and algebraic geometry. Med Image Anal. 2021;70:101994.
78. 3dStool. Available from: https://github.com/SpyrosSou/3dStool. [Last accessed on 2 Jul 2024].
79. Greene N, Luo W, Kazanzides P. dvpose: automated data collection and dataset for 6d pose estimation of robotic surgical instruments. In: 2023 International Symposium on Medical Robotics (ISMR); 2023 Apr 19-21; Atlanta, GA, USA. IEEE; 2023. pp. 1-7.
80. Fisher R. Edinburgh simulated surgical tools dataset (RGBD). 2022. Available from: https://groups.inf.ed.ac.uk/vision/DATASETS/SURGICALTOOLS/. [Last accessed on 2 Jul 2024].
81. 6-dof pose estimation of surgical instruments. 2022. Available from: https://www.kaggle.com/datasets/juanantoniobarragan/6-dof-pose-estimation-of-surgical-instruments. [Last accessed on 2 Jul 2024].
82. Munawar A, Wu JY, Fischer GS, Taylor RH, Kazanzides P. Open simulation environment for learning and practice of robot-assisted surgical suturing. IEEE Robot Autom Lett. 2022;7:3843-50.
83. Wang R, Ktistakis S, Zhang S, Meboldt M, Lohmeyer Q. POV-surgery: a dataset for egocentric hand and tool pose estimation during surgical activities. In: Greenspan H, et al., editors. Medical Image Computing and Computer Assisted Intervention - MICCAI 2023. Cham: Springer; 2023. pp. 440-50.
84. Kügler D, Sehring J, Stefanov A, et al. i3PosNet: instrument pose estimation from X-ray in temporal bone surgery. Int J Comput Assist Radiol Surg. 2020;15:1137-45.
85. Zhang J, Hu J. Image segmentation based on 2d otsu method with histogram analysis. In: 2008 International Conference on Computer Science and Software Engineering; 2008 Dec 12-14; Wuhan, China. IEEE; 2008. pp. 105-8.
86. Pham DL, Prince JL. An adaptive fuzzy C-means algorithm for image segmentation in the presence of intensity inhomogeneities. Pattern Recognit Lett. 1999;20:57-68.
87. Lin C, Chen C. Image segmentation based on edge detection and region growing for thinprep-cervical smear. Int J Patt Recogn Artif Intell. 2010;24:1061-89.
88. Dalal N, Triggs B. Histograms of oriented gradients for human detection. In: 2005 IEEE Computer Society Conference on Computer Vision and Pattern Recognition (CVPR’05); 2005 Jun 20-25; San Diego, CA, USA. IEEE; 2005. pp. 886-93.
89. Felzenszwalb PF, Girshick RB, McAllester D, Ramanan D. Object detection with discriminatively trained part-based models. IEEE Trans Pattern Anal Mach Intell. 2010;32:1627-45.
90. Viola P, Jones M. Rapid object detection using a boosted cascade of simple features. In: Proceedings of the 2001 IEEE Computer Society Conference on Computer Vision and Pattern Recognition. CVPR 2001; 2001 Dec 8-14; Kauai, HI, USA. IEEE; 2001. p. 1.
91. Long J, Shelhamer E, Darrell T. Fully convolutional networks for semantic segmentation. In: 2015 IEEE Conference on Computer Vision and Pattern Recognition (CVPR). IEEE; 2015. pp. 3431-40.
92. Ronneberger O, Fischer P, Brox T. U-Net: convolutional networks for biomedical image segmentation. In: Navab N, Hornegger J, Wells WM, Frangi AF, editors. Medical Image Computing and Computer-Assisted Intervention - MICCAI 2015. Cham: Springer; 2015. pp. 234-41.
93. Chen L, Zhu Y, Papandreou G, Schroff F, Adam H. Encoder-decoder with atrous separable convolution for semantic image segmentation. In: Ferrari V, Hebert M, Sminchisescu C, Weiss Y, editors. Computer Vision - ECCV 2018. Cham: Springer; 2018. pp. 833-51.
94. Zhao H, Shi J, Qi X, Wang X, Jia J. Pyramid scene parsing network. In: 2017 IEEE Conference on Computer Vision and Pattern Recognition (CVPR); 2017 Jul 21-26; Honolulu, HI, USA. IEEE; 2017. pp. 2881-90.
95. Seenivasan L, Mitheran S, Islam M, Ren H. Global-reasoned multi-task learning model for surgical scene understanding. IEEE Robot Autom Lett. 2022;7:3858-65.
96. Girshick R. Fast r-cnn. In: 2015 IEEE International Conference on Computer Vision (ICCV); 2015 Dec 7-13; Santiago, Chile. IEEE; 2015. pp. 1440-8.
97. Ren S, He K, Girshick R, Sun J. Faster r-cnn: towards real-time object detection with region proposal networks. Available from: https://proceedings.neurips.cc/paper/2015/hash/14bfa6bb14875e45bba028a21ed38046-Abstract.html. [Last accessed on 2 Jul 2024]
98. Girshick R, Donahue J, Darrell T, Malik J. Rich feature hierarchies for accurate object detection and semantic segmentation. In: 2014 IEEE Conference on Computer Vision and Pattern Recognition; 2014 Jun 23-28; Columbus, OH, USA. IEEE; 2014. pp. 580-7.
99. Redmon J, Divvala S, Girshick R, Farhadi A. You only look once: unified, real-time object detection. In: 2016 IEEE Conference on Computer Vision and Pattern Recognition (CVPR); 2016 Jun 27-30; Las Vegas, NV, USA. IEEE; 2016. pp. 779-88.
100. Liu W, Anguelov D, Erhan D, et al. SSD: single shot multibox detector. In: Leibe B, Matas J, Sebe N, Welling M, editors. Computer Vision - ECCV 2016. Cham: Springer; 2016. pp. 21-37.
101. Lu X, Li B, Yue Y, Li Q, Yan J. Grid R-CNN. In: 2019 IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR); 2019 Jun 15-20; Long Beach, CA, USA. IEEE; 2019. pp. 7363-72.
102. Zhang H, Chang H, Ma B, Wang N, Chen X. Dynamic R-CNN: towards high quality object detection via dynamic training. In: Vedaldi A, Bischof H, Brox T, Frahm J, editors. Computer Vision - ECCV 2020. Cham: Springer; 2020. pp. 260-75.
103. Wu Y, Chen Y, Yuan L, et al. Rethinking classification and localization for object detection. In: 2020 IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR); 2020 Jun 13-19; Seattle, WA, USA. IEEE; 2020. pp. 10186-92.
104. Lin TY, Goyal P, Girshick R, He K, Dollár P. Focal loss for dense object detection. In: 2017 IEEE International Conference on Computer Vision (ICCV); 2017 Oct 22-29; Venice, Italy. IEEE; 2017. pp. 2980-8.
105. Law H, Deng J. CornerNet: detecting objects as paired keypoints. In: Ferrari V, Hebert M, Sminchisescu C, Weiss Y, editors. Computer Vision - ECCV 2018. Cham: Springer; 2018. pp. 765-81.
106. Zhou X, Wang D, Krähenbühl P. Objects as points. arXiv. [Preprint.] Apr 16, 2019 [accessed 2024 Jul 3]. Available from: https://arxiv.org/abs/1904.07850.
107. Yang Z, Liu S, Hu H, Wang L, Lin S. Reppoints: point set representation for object detection. In: 2019 IEEE/CVF International Conference on Computer Vision (ICCV); 2019 Oct 27 - Nov 2; Seoul, Korea (South). IEEE; 2019. pp. 9657-66.
108. Tian Z, Shen C, Chen H, He T. FCOS: a simple and strong anchor-free object detector. IEEE Trans Pattern Anal Mach Intell. 2022;44:1922-33.
109. He K, Gkioxari G, Dollár P, Girshick R. Mask R-CNN. In: 2017 IEEE International Conference on Computer Vision (ICCV); 2017 Oct 22-29; Venice, Italy. IEEE; 2017. pp. 2980-8.
110. Huang Z, Huang L, Gong Y, Huang C, Wang X. Mask scoring R-CNN. In: 2019 IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR); 2019 Jun 15-20; Long Beach, CA, USA. IEEE; 2019. pp. 6402-11.
111. Chen K, Pang J, Wang J, et al. Hybrid task cascade for instance segmentation. In: 2019 IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR); 2019 Jun 15-20; Long Beach, CA, USA. IEEE; 2019. pp. 4969-78.
112. Ding H, Qiao S, Yuille A, Shen W. Deeply shape-guided cascade for instance segmentation. In: 2021 IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR); 2021 Jun 20-25; Nashville, TN, USA. IEEE; 2021. pp. 8274-84.
113. Bolya D, Zhou C, Xiao F, Lee YJ. Yolact: real-time instance segmentation. In: 2019 IEEE/CVF International Conference on Computer Vision (ICCV); 2019 Oct 27 - Nov 2; Seoul, Korea (South). IEEE; 2019. pp. 9156-65.
114. Wang X, Kong T, Shen C, Jiang Y, Li L. SOLO: segmenting objects by locations. In: Vedaldi A, Bischof H, Brox T, Frahm J, editors. Computer Vision - ECCV 2020. Cham: Springer; 2020. pp. 649-65.
115. Kirillov A, Wu Y, He K, Girshick R. Pointrend: image segmentation as rendering. In: 2020 IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR); 2020 Jun 13-19; Seattle, WA, USA. IEEE; 2020. pp. 9796-805.
116. Tian Z, Shen C, Chen H. Conditional convolutions for instance segmentation. In: Vedaldi A, Bischof H, Brox T, Frahm J, editors. Computer Vision - ECCV 2020. Cham: Springer; 2020. pp. 282-98.
117. Liu K, Zhao Z, Shi P, Li F, Song H. Real-time surgical tool detection in computer-aided surgery based on enhanced feature-fusion convolutional neural network. J Comput Des Eng. 2022;9:1123-34.
118. Bamba Y, Ogawa S, Itabashi M, et al. Object and anatomical feature recognition in surgical video images based on a convolutional neural network. Int J Comput Assist Radiol Surg. 2021;16:2045-54.
119. Cerón JCÁ, Ruiz GO, Chang L, Ali S. Real-time instance segmentation of surgical instruments using attention and multi-scale feature fusion. Med Image Anal. 2022;81:102569.
120. Wang A, Islam M, Xu M, Ren H. Rethinking surgical instrument segmentation: a background image can be all you need. In: Wang L, Dou Q, Fletcher PT, Speidel S, Li S, editors. Medical Image Computing and Computer Assisted Intervention - MICCAI 2022. Cham: Springer; 2022. pp. 355-64.
121. Zhang Z, Rosa B, Nageotte F. Surgical tool segmentation using generative adversarial networks with unpaired training data. IEEE Robot Autom Lett. 2021;6:6266-73.
122. Yang L, Gu Y, Bian G, Liu Y. An attention-guided network for surgical instrument segmentation from endoscopic images. Comput Biol Med. 2022;151:106216.
123. Ding H, Wu JY, Li Z, Unberath M. Rethinking causality-driven robot tool segmentation with temporal constraints. Int J Comput Assist Radiol Surg. 2023;18:1009-16.
124. Colleoni E, Edwards P, Stoyanov D. Synthetic and real inputs for tool segmentation in robotic surgery. In: Martel AL, et al., editors. Medical Image Computing and Computer Assisted Intervention - MICCAI 2020. Cham: Springer; 2020. pp. 700-10.
125. Lee K, Choi MK, Jung H. DavinciGAN: unpaired surgical instrument translation for data augmentation. Available from: http://proceedings.mlr.press/v102/lee19a.html. [Last accessed on 3 Jul 2024]
126. Zheng S, Lu J, Zhao H, et al. Rethinking semantic segmentation from a sequence-to-sequence perspective with transformers. In: 2021 IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR); 2021 Jun 20-25; Nashville, TN, USA. IEEE; 2021. pp. 6877-86.
127. Xie E, Wang W, Yu Z, Anandkumar A, Alvarez JM, Luo P. Segformer: simple and efficient design for semantic segmentation with transformers. In: Advances in neural information processing systems 34 (NeurIPS 2021). Available from: https://proceedings.neurips.cc/paper/2021/hash/64f1f27bf1b4ec22924fd0acb550c235-Abstract.html. [Last accessed on 3 Jul 2024]
128. Carion N, Massa F, Synnaeve G, Usunier N, Kirillov A, Zagoruyko S. End-to-end object detection with transformers. In: Vedaldi A, Bischof H, Brox T, Frahm J, editors. Computer Vision - ECCV 2020. Cham: Springer; 2020. pp. 213-29.
129. Zhu X, Su W, Lu L, Li B, Wang X, Dai J. Deformable detr: deformable transformers for end-to-end object detection. arXiv. [Preprint.] Mar 18, 2021 [accessed 2024 Jul 3]. Available from: https://arxiv.org/abs/2010.04159.
130. Meng D, Chen X, Fan Z, et al. Conditional DETR for fast training convergence. In: 2021 IEEE/CVF International Conference on Computer Vision (ICCV); 2021 Oct 10-17; Montreal, QC, Canada. IEEE; 2021. pp. 3631-40.
131. Li Y, Mao H, Girshick R, He K. Exploring plain vision transformer backbones for object detection. In: Avidan S, Brostow G, Cissé M, Farinella GM, Hassner T, editors. Computer Vision - ECCV 2022. Cham: Springer; 2022. pp. 280-96.
132. Zhang H, Li F, Liu S, et al. DINO: DETR with improved denoising anchor boxes for end-to-end object detection. arXiv. [Preprint.] Jul 11, 2022 [accessed 2024 Jul 3]. Available from: https://arxiv.org/abs/2203.03605.
133. Cheng B, Misra I, Schwing AG, Kirillov A, Girdhar R. Masked-attention mask transformer for universal image segmentation. In: 2022 IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR); 2022 Jun 18-24; New Orleans, LA, USA. IEEE; 2022. pp. 1280-9.
134. Zou X, Dou ZY, Yang J, et al. Generalized decoding for pixel, image, and language. In: 2023 IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR); 2023 Jun 17-24; Vancouver, BC, Canada. IEEE; 2023. pp. 15116-27.
135. Radford A, Kim JW, Hallacy C, et al. Learning transferable visual models from natural language supervision. Available from: http://proceedings.mlr.press/v139/radford21a.html. [Last accessed on 3 Jul 2024]
136. Li LH, Zhang P, Zhang H, et al. Grounded language-image pre-training. In: 2022 IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR); 2022 Jun 18-24; New Orleans, LA, USA. IEEE; 2022. pp. 10955-65.
137. Zhong Y, Yang J, Zhang P, et al. RegionCLIP: region-based language-image pretraining. In: 2022 IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR); 2022 Jun 18-24; New Orleans, LA, USA. IEEE; 2022. 16772-82.
138. Luo H, Bao J, Wu Y, He X, Li T. SegCLIP: patch aggregation with learnable centers for open-vocabulary semantic segmentation. Available from: https://proceedings.mlr.press/v202/luo23a.html. [Last accessed on 3 Jul 2024]
139. He Z, Unberath M, Ke J, Shen Y. TransNuSeg: a lightweight multi-task transformer for nuclei segmentation. In: Greenspan H, et al., editors. Medical Image Computing and Computer Assisted Intervention - MICCAI 2023. Cham: Springer; 2023. pp. 206-15.
140. Shen Y, Guo P, Wu J, et al. MoViT: Memorizing vision transformers for medical image analysis. In: Cao X, Xu X, Rekik I, Cui Z, Ouyang X, editors. Machine Learning in Medical Imaging. Cham: Springer; 2024. pp. 205-13.
141. Oguine K, Soberanis-Muku R, Drenkow N, Unberath M. From generalization to precision: exploring sam for tool segmentation in surgical environments. Med Imag Proc 2024 Imag Process. 2024;12926:7-12.
142. Peng Z, Xu Z, Zeng Z, Yang X, Shen W. SAM-PARSER: fine-tuning SAM efficiently by parameter space reconstruction. arXiv. [Preprint.] Dec 18, 2023 [accessed 2024 Jul 3]. Available from: https://arxiv.org/abs/2308.14604.
143. Li X, Zhang Y, Zhao L. Multi-prompt fine-tuning of foundation models for enhanced medical image segmentation. arXiv. [Preprint.] Oct 3, 2023 [accessed 2024 Jul 3]. Available from: https://arxiv.org/abs/2310.02381.
144. Tyagi AK, Mishra V, Prathosh AP, Mausam. Guided prompting in sam for weakly supervised cell segmentation in histopathological images. arXiv. [Preprint.] Nov 29, 2023 [accessed 2024 Jul 3]. Available from: https://arxiv.org/abs/2311.17960.
145. Paranjape JN, Nair NG, Sikder S, Vedula SS, Patel VM. AdaptiveSAM: towards efficient tuning of SAM for surgical scene segmentation. arXiv. [Preprint.] Aug 7, 2023 [accessed 2024 Jul 3]. Available from: https://arxiv.org/abs/2308.03726.
146. Yue W, Zhang J, Hu K, Xia Y, Luo J, Wang Z. SurgicalSAM: efficient class promptable surgical instrument segmentation. arXiv. [Preprint.] Dec 21, 2023 [accessed 2024 Jul 3]. Available from: https://arxiv.org/abs/2308.08746.
147. Wang A, Islam M, Xu M, Zhang Y, Ren H. SAM meets robotic surgery: an empirical study in robustness perspective. arXiv. [Preprint.] Apr 28, 2023 [accessed 2024 Jul 3]. Available from: https://arxiv.org/abs/2304.14674.
148. He Y, Yu H, Liu X, Yang Z, Sun W, Mian A. Deep learning based 3D segmentation: a survey. arXiv. [Preprint.] Jul 26, 2023 [accessed 2024 Jul 3]. Available from: https://arxiv.org/abs/2103.05423.
149. Qian R, Lai X, Li X. 3D object detection for autonomous driving: a survey. Pattern Recognit. 2022;130:108796.
150. Qi CR, Su H, Mo K, Guibas LJ. Pointnet: deep learning on point sets for 3D classification and segmentation. In: 2017 IEEE Conference on Computer Vision and Pattern Recognition (CVPR); 2017 Jul 21-26; Honolulu, HI, USA. IEEE; 2017. pp. 77-85.
151. Wang W, Neumann U. Depth-aware CNN for RGB-D segmentation. In: Ferrari V, Hebert M, Sminchisescu C, Weiss Y, editors. Computer Vision - ECCV 2018. Cham: Springer; 2018. pp. 144-61.
152. Zhang Y, Lu J, Zhou J. Objects are different: flexible monocular 3d object detection. In: 2021 IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR); 2021 Jun 20-25; Nashville, TN, USA. IEEE; 2021. pp. 3288-97.
153. Wang Y, Guizilini VC, Zhang T, Wang Y, Zhao H, Solomon J. DETR3D: 3D object detection from multi-view images via 3D-to-2D queries. Available from: https://proceedings.mlr.press/v164/wang22b.html. [Last accessed on 3 Jul 2024]
154. Maninis KK, Caelles S, Chen Y, et al. Video object segmentation without temporal information. IEEE Trans Pattern Anal Mach Intell. 2019;41:1515-30.
155. Caelles S, Maninis KK, Pont-Tuset J, Leal-Taixé L, Cremers D, Van Gool L. One-shot video object segmentation. In: 2017 IEEE Conference on Computer Vision and Pattern Recognition (CVPR); 2017 Jul 21-26; Honolulu, HI, USA. IEEE; 2017. pp. 221-30.
156. Hu YT, Huang JB, Schwing A. MaskRNN: instance level video object segmentation. Available from: https://proceedings.neurips.cc/paper/2017/hash/6c9882bbac1c7093bd25041881277658-Abstract.html. [Last accessed on 3 Jul 2024]
157. Ventura C, Bellver M, Girbau A, Salvador A, Marques F, Giro-i-Nieto X. RVOS: end-to-end recurrent network for video object segmentation. In: 2019 IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR); Jun 15-20; Long Beach, CA, USA. IEEE; 2019. pp. 5272-81.
158. Oh SW, Lee JY, Xu N, Kim SJ. Video object segmentation using space-time memory networks. In: 2019 IEEE/CVF International Conference on Computer Vision (ICCV); 2019 Oct 27 - Nov 2; Seoul, Korea (South). IEEE; 2019. pp. 9225-34.
159. Cheng HK, Tai YW, Tang CK. Rethinking space-time networks with improved memory coverage for efficient video object segmentation. Available from: https://proceedings.neurips.cc/paper/2021/hash/61b4a64be663682e8cb037d9719ad8cd-Abstract.html. [Last accessed on 3 Jul 2024]
160. Cheng HK, Schwing AG. XMem: long-term video object segmentation with an atkinson-shiffrin memory model. In: Avidan S, Brostow G, Cissé M, Farinella GM, Hassner T, editors. Computer Vision - ECCV 2022. Cham: Springer; 2022. pp. 640-58.
161. Duke B, Ahmed A, Wolf C, Aarabi P, Taylor GW. SSTVOS: sparse spatiotemporal transformers for video object segmentation. In: 2021 IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR); 2021 Jun 20-25; Nashville, TN, USA. IEEE; 2021. pp. 5908-17.
162. Yang Z, Wei Y, Yang Y. Associating objects with transformers for video object segmentation. Available from: https://proceedings.neurips.cc/paper/2021/hash/147702db07145348245dc5a2f2fe5683-Abstract.html. [Last accessed on 3 Jul 2024]
163. Cheng HK, Oh SW, Price B, Lee JY, Schwing A. Putting the object back into video object segmentation. arXiv. [Preprint.] Apr 11, 2024 [accessed 2024 Jul 3]. Available from: https://arxiv.org/abs/2310.12982.
164. Gong T, Chen K, Wang X, et al. Temporal ROI align for video object recognition. AAAI. 2021;35:1442-50.
165. Wu H, Chen Y, Wang N, Zhang ZX. Sequence level semantics aggregation for video object detection. In: 2019 IEEE/CVF International Conference on Computer Vision (ICCV); 2019 Oct 27 - Nov 2. IEEE; 2019. pp. 9216-24.
166. Zhu X, Wang Y, Dai J, Yuan L, Wei Y. Flow-guided feature aggregation for video object detection. In: 2017 IEEE International Conference on Computer Vision (ICCV); 2017 Oct 22-29; Venice, Italy. IEEE; 2017. pp. 408-17.
167. Zhu X, Xiong Y, Dai J, Yuan L, Wei Y. Deep feature flow for video recognition. In: 2017 IEEE Conference on Computer Vision and Pattern Recognition (CVPR). IEEE; 2017. pp. 4141-50.
168. Li B, Wu W, Wang Q, Zhang F, Xing J, Yan J. SiamRPN++: evolution of siamese visual tracking with very deep networks. In: 2019 IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR); 2019 Jun 15-20; Long Beach, CA, USA. IEEE; 2019. pp. 4277-86.
169. Yan B, Peng H, Fu J, Wang D, Lu H. Learning spatio-temporal transformer for visual tracking. In: 2021 IEEE/CVF International Conference on Computer Vision (ICCV); 2021 Oct 10-17; Montreal, QC, Canada. IEEE; 2021. pp. 10428-37.
170. Cui Y, Jiang C, Wang L, Wu G. Mixformer: end-to-end tracking with iterative mixed attention. In: 2022 IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR); 2022 JUN 18-24; New Orleans, LA, USA. IEEE; 2022. pp. 13598-608.
171. Bergmann P, Meinhardt T, Leal-Taixe L. Tracking without bells and whistles. In: 2019 IEEE/CVF International Conference on Computer Vision (ICCV); 2019 Oct 27 - Nov 2; Seoul, Korea (South). IEEE; 2019. pp. 941-51.
172. Pang J, Qiu L, Li X, et al. Quasi-dense similarity learning for multiple object tracking. In: 2021 IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR); 2021 Jun 20-25; Nashville, TN, USA. IEEE; 2021. pp. 164-73.
173. Zhang Y, Sun P, Jiang Y, et al. ByteTrack: Multi-object tracking by associating every detection box. In: Avidan S, Brostow G, Cissé M, Farinella GM, Hassner T, editors. Computer Vision - ECCV 2022. Cham: Springer; 2022. pp. 1-21.
174. Luo W, Xing J, Milan A, Zhang X, Liu W, Kim T. Multiple object tracking: a literature review. Artif Intell. 2021;293:103448.
175. Wang Y, Sun Q, Liu Z, Gu L. Visual detection and tracking algorithms for minimally invasive surgical instruments: a comprehensive review of the state-of-the-art. Robot Auton Syst. 2022;149:103945.
176. Dakua SP, Abinahed J, Zakaria A, et al. Moving object tracking in clinical scenarios: application to cardiac surgery and cerebral aneurysm clipping. Int J Comput Assist Radiol Surg. 2019;14:2165-76.
177. Liu B, Sun M, Liu Q, Kassam A, Li CC, Sclabassi R. Automatic detection of region of interest based on object tracking in neurosurgical video. Conf Proc IEEE Eng Med Biol Soc. 2005;2005:6273-6.
178. Du X, Allan M, Bodenstedt S, et al. Patch-based adaptive weighting with segmentation and scale (PAWSS) for visual tracking in surgical video. Med Image Anal. 2019;57:120-35.
179. Stenmark M, Omerbašić E, Magnusson M, Andersson V, Abrahamsson M, Tran PK. Vision-based tracking of surgical motion during live open-heart surgery. J Surg Res. 2022;271:106-16.
180. Cheng T, Li W, Ng WY, et al. Deep learning assisted robotic magnetic anchored and guided endoscope for real-time instrument tracking. IEEE Robot Autom Lett. 2021;6:3979-86.
181. Zhao Z, Voros S, Chen Z, Cheng X. Surgical tool tracking based on two CNNs: from coarse to fine. J Eng. 2019;2019:467-72.
182. Robu M, Kadkhodamohammadi A, Luengo I, Stoyanov D. Towards real-time multiple surgical tool tracking. Comput Methods Biomecha Biomed Eng Imaging Vis. 2021;9:279-85.
183. Lee D, Yu HW, Kwon H, Kong HJ, Lee KE, Kim HC. Evaluation of surgical skills during robotic surgery by deep learning-based multiple surgical instrument tracking in training and actual operations. J Clin Med. 2020;9:1964.
184. García-Peraza-Herrera LC, Li W, Gruijthuijsen C, et al. Real-time segmentation of non-rigid surgical tools based on deep learning and tracking. In: Peters T, et al., editors. Computer-Assisted and Robotic Endoscopy. Cham: Springer; 2017. pp. 84-95.
185. Jo K, Choi Y, Choi J, Chung JW. Robust real-time detection of laparoscopic instruments in robot surgery using convolutional neural networks with motion vector prediction. Appl Sci. 2019;9:2865.
186. Zhao Z, Chen Z, Voros S, Cheng X. Real-time tracking of surgical instruments based on spatio-temporal context and deep learning. Comput Assist Surg. 2019;24:20-9.
187. Alshirbaji TA, Jalal NA, Möller K. A convolutional neural network with a two-stage LSTM model for tool presence detection in laparoscopic videos. Curr Dir Biomed Eng. 2020;6:20200002.
188. Bouguet JY, Perona P. 3D photography using shadows in dual-space geometry. Int J Comput Vis. 1999;35:129-49.
189. Iddan GJ, Yahav G. Three-dimensional imaging in the studio and elsewhere. Proc SPIE. 2001;4298:48-55.
190. Nayar SK, Krishnan G, Grossberg MD, Raskar R. Fast separation of direct and global components of a scene using high frequency illumination. ACM Trans Graph. 2006;25:935-44.
191. Torralba A, Oliva A. Depth estimation from image structure. IEEE Trans Pattern Anal Mach Intell. 2002;24:1226-38.
192. Marr D, Poggio T. Cooperative computation of stereo disparity: a cooperative algorithm is derived for extracting disparity information from stereo image pairs. Science. 1976;194:283-7.
194. Hannah MJ. Computer matching of areas in stereo images. Stanford University. 1974. Available from: https://www.semanticscholar.org/paper/Computer-matching-of-areas-in-stereo-images.-Hannah/02a0829a658e7dbfdf49e8112b38f8911a12eb76. [Last accessed on 3 Jul 2024].
195. Stoyanov D, Darzi A, Yang GZ. A practical approach towards accurate dense 3D depth recovery for robotic laparoscopic surgery. Comput Aided Surg. 2005;10:199-208.
196. Arnold RD. Automated stereo perception. PhD book, Stanford University (1983). Available from: https://searchworks.stanford.edu/view/1052936. [Last accessed on 3 Jul 2024]
197. Okutomi M, Kanade T. A locally adaptive window for signal matching. Int J Comput Vision. 1992;7:143-62.
199. Lo B, Scarzanella MV, Stoyanov D, Yang G. Belief propagation for depth cue fusion in minimally invasive surgery. In: Metaxas D, Axel L, Fichtinger G, Székely G, editors. Medical Image Computing and Computer-Assisted Intervention - MICCAI 2008. Berlin: Springer; 2008. pp. 104-12.
200. Sinha RY, Raje SR, Rao GA. Three-dimensional laparoscopy: principles and practice. J Minim Access Surg. 2017;13:165-9.
201. Mueller-Richter UD, Limberger A, Weber P, Ruprecht KW, Spitzer W, Schilling M. Possibilities and limitations of current stereo-endoscopy. Surg Endosc. 2004;18:942-7.
202. Bogdanova R, Boulanger P, Zheng B. Depth perception of surgeons in minimally invasive surgery. Surg Innov. 2016;23:515-24.
203. Sinha R, Sundaram M, Raje S, Rao G, Sinha M, Sinha R. 3D laparoscopy: technique and initial experience in 451 cases. Gynecol Surg. 2013;10:123-8.
204. Liu X, Sinha A, Ishii M, et al. Dense depth estimation in monocular endoscopy with self-supervised learning methods. IEEE Trans Med Imaging. 2020;39:1438-47.
205. Li L, Li X, Yang S, Ding S, Jolfaei A, Zheng X. Unsupervised-learning-based continuous depth and motion estimation with monocular endoscopy for virtual reality minimally invasive surgery. IEEE Trans Ind Inf. 2021;17:3920-8.
206. Liu F, Shen C, Lin G. Deep convolutional neural fields for depth estimation from a single image. In: 2015 IEEE Conference on Computer Vision and Pattern Recognition (CVPR); 2015 Jun 7-12; Boston, MA, USA. IEEE; 2014. pp. 5162-70.
207. Visentini-Scarzanella M, Sugiura T, Kaneko T, Koto S. Deep monocular 3D reconstruction for assisted navigation in bronchoscopy. Int J Comput Assist Radiol Surg. 2017;12:1089-99.
208. Oda M, Itoh H, Tanaka K, et al. Depth estimation from single-shot monocular endoscope image using image domain adaptation and edge-aware depth estimation. Comput Methods Biomech Biomed Eng Imaging Vis. 2022;10:266-73.
209. Zhan H, Garg R, Weerasekera CS, Li K, Agarwal H, Reid I. Unsupervised learning of monocular depth estimation and visual odometry with deep feature reconstruction. In: 2018 IEEE/CVF Conference on Computer Vision and Pattern Recognition; 2018 Jun 18-23; Salt Lake City, UT, USA. IEEE; 2018. pp. 340-9.
210. Liu F, Jonmohamadi Y, Maicas G, Pandey AK, Carneiro G. Self-supervised depth estimation to regularise semantic segmentation in knee arthroscopy. In: Martel AL, et al., editors. Medical Image Computing and Computer Assisted Intervention - MICCAI 2020. Cham: Springer; 2020. pp. 594-603.
211. Mahmood F, Chen R, Durr NJ. Unsupervised reverse domain adaptation for synthetic medical images via adversarial training. IEEE Trans Med Imaging. 2018;37:2572-81.
212. Guo R, Ayinde B, Sun H, Muralidharan H, Oguchi K. Monocular depth estimation using synthetic images with shadow removal. In: 2019 IEEE Intelligent Transportation Systems Conference (ITSC); 2019 Oct 27-30; Auckland, New Zealand. IEEE; 2019. pp. 1432-9.
213. Chen RJ, Bobrow TL, Athe T, Mahmood F, Durr NJ. SLAM endoscopy enhanced by adversarial depth prediction. arXiv. [Preprint.] Jun 29, 2019 [accessed 2024 Jul 3]. Available from: https://arxiv.org/abs/1907.00283.
214. Schreiber AM, Hong M, Rozenblit JW. Monocular depth estimation using synthetic data for an augmented reality training system in laparoscopic surgery. In: 2021 IEEE International Conference on Systems, Man, and Cybernetics (SMC); 2021 Oct 17-20; Melbourne, Australia. IEEE; 2021. pp. 2121-6.
215. Tong HS, Ng YL, Liu Z, et al. Real-to-virtual domain transfer-based depth estimation for real-time 3D annotation in transnasal surgery: a study of annotation accuracy and stability. Int J Comput Assist Radiol Surg. 2021;16:731-9.
216. Wong A, Soatto S. Bilateral cyclic constraint and adaptive regularization for unsupervised monocular depth prediction. In: 2019 IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR); 2019 Jun 15-20; Long Beach, CA, USA. IEEE; 2019. pp. 5637-46.
217. Widya AR, Monno Y, Okutomi M, Suzuki S, Gotoda T, Miki K. Learning-based depth and pose estimation for monocular endoscope with loss generalization. Annu Int Conf IEEE Eng Med Biol Soc. 2021;2021:3547-52.
218. Shao S, Pei Z, Chen W, et al. Self-Supervised monocular depth and ego-Motion estimation in endoscopy: appearance flow to the rescue. Med Image Anal. 2022;77:102338.
219. Hwang SJ, Park SJ, Kim GM, Baek JH. Unsupervised monocular depth estimation for colonoscope system using feedback network. Sensors. 2021;21:2691.
220. Li W, Hayashi Y, Oda M, Kitasaka T, Misawa K, Mori K. Geometric constraints for self-supervised monocular depth estimation on laparoscopic images with dual-task consistency. In: Wang L, Dou Q, Fletcher PT, Speidel S, Li S, editors. Medical Image Computing and Computer Assisted Intervention - MICCAI 2022. Cham: Springer; 2022. pp. 467-77.
221. Masuda T, Sagawa R, Furukawa R, Kawasaki H. Scale-preserving shape reconstruction from monocular endoscope image sequences by supervised depth learning. Healthc Technol Lett. 2024;11:76-84.
222. Tukra S, Giannarou S. Randomly connected neural networks for self-supervised monocular depth estimation. Comput Methods Biomech Biomed Eng Imaging Vis. 2022;10:390-9.
223. Zhao S, Wang C, Wang Q, Liu Y, Zhou SK. 3D endoscopic depth estimation using 3d surface-aware constraints. arXiv. [Preprint.] Mar 4, 2022 [accessed 2024 Jul 3]. Available from: https://arxiv.org/abs/2203.02131.
224. Han J, Jiang Z, Feng G. Monocular depth estimation based on chained residual pooling and gradient weighted loss. In: 2023 3rd International Conference on Consumer Electronics and Computer Engineering (ICCECE); 2023 Jan 6-8; Guangzhou, China. IEEE; 2023. pp. 278-82.
225. Yang Y, Shao S, Yang T, et al. A geometry-aware deep network for depth estimation in monocular endoscopy. Eng Appl Artif Intell. 2023;122:105989.
226. Zhang G, Gao X, Meng H, Pang Y, Nie X. A self-supervised network-based smoke removal and depth estimation for monocular endoscopic videos. IEEE Trans Vis Comput Graph. 2024;30:6547-59.
227. Yang L, Kang B, Huang Z, Xu X, Feng J, Zhao H. Depth anything: unleashing the power of large-scale unlabeled data. arXiv. [Preprint.] Apr 7, 2024 [accessed 2024 Jul 3]. Available from: https://arxiv.org/abs/2401.10891.
228. Han JJ, Acar A, Henry C, Wu JY. Depth anything in medical images: a comparative study. arXiv. [Preprint.] Jan 29, 2024 [accessed 2024 Jul 3]. Available from: https://arxiv.org/abs/2401.16600.
229. Chang JR, Chen YS. Pyramid stereo matching network. In: 2018 IEEE/CVF Conference on Computer Vision and Pattern Recognition; 2018 Jun 18-23; Salt Lake City, UT, USA. IEEE; 2018. pp. 5410-8.
230. Luo W, Schwing AG, Urtasun R. Efficient deep learning for stereo matching. In: 2016 IEEE Conference on Computer Vision and Pattern Recognition (CVPR); 2016 Jun 27-30; Las Vegas, NV, USA. IEEE; 2016. pp. 5695-703.
231. Zampokas G, Peleka G, Tsiolis K, Topalidou-Kyniazopoulou A, Mariolis I, Tzovaras D. Real-time stereo reconstruction of intraoperative scene and registration to preoperative 3D models for augmenting surgeons’ view during RAMIS. Med Phys. 2022;49:6517-26.
232. Probst T, Maninis K, Chhatkuli A, Ourak M, Poorten EV, Van Gool L. Automatic tool landmark detection for stereo vision in robot-assisted retinal surgery. IEEE Robot Autom Lett. 2018;3:612-9.
233. Dosovitskiy A, Beyer L, Kolesnikov A, et al. An image is worth 16x16 words: transformers for image recognition at scale. arXiv. [Preprint.] Jun 3, 2021 [accessed 2024 Jul 3]. Available from: https://arxiv.org/abs/2010.11929.
234. Tao R, Huang B, Zou X, Zheng G. SVT-SDE: spatiotemporal vision transformers-based self-supervised depth estimation in stereoscopic surgical videos. IEEE Trans Med Robot Bionics. 2023;5:42-53.
235. Li Z, Liu X, Drenkow N, et al. Revisiting stereo depth estimation from a sequence-to-sequence perspective with transformers. In: 2021 IEEE/CVF International Conference on Computer Vision (ICCV); 2021 Oct 10-17; Montreal, QC, Canada. IEEE; 2021. pp. 6177-86.
236. Long Y, Li Z, Yee CH, et al. E-DSSR: efficient dynamic surgical scene reconstruction with transformer-based stereoscopic depth perception. In: de Bruijne M, et al., editors. Medical Image Computing and Computer Assisted Intervention - MICCAI 2021. Cham: Springer; 2021. pp. 415-25.
237. Guo W, Li Z, Yang Y, et al. Context-enhanced stereo transformer. In: Avidan S, Brostow G, Cissé M, Farinella GM, Hassner T, editors. Computer Vision - ECCV 2022. Cham: Springer; 2022. pp. 263-79.
238. Hu X, Baena FRY. Automatic bone surface restoration for markerless computer-assisted orthopaedic surgery. Chin J Mech Eng. 2022;35:18.
239. Wang W, Zhou H, Yan Y, et al. An automatic extraction method on medical feature points based on PointNet++ for robot-assisted knee arthroplasty. Int J Med Robot. 2023;19:e2464.
240. Baum ZMC, Hu Y, Barratt DC. Multimodality biomedical image registration using free point transformer networks. In: Hu Y, et al., editors. Medical Ultrasound, and Preterm, Perinatal and Paediatric Image Analysis. Cham: Springer; 2020. pp. 116-25.
241. Baum ZMC, Hu Y, Barratt DC. Real-time multimodal image registration with partial intraoperative point-set data. Med Image Anal. 2021;74:102231.
242. Widya AR, Monno Y, Imahori K, et al. 3D reconstruction of whole stomach from endoscope video using structure-from-motion. Annu Int Conf IEEE Eng Med Biol Soc. 2019;2019:3900-4.
243. Lin B, Sun Y, Qian X, Goldgof D, Gitlin R, You Y. Video-based 3D reconstruction, laparoscope localization and deformation recovery for abdominal minimally invasive surgery: a survey. Int J Med Robot. 2016;12:158-78.
244. Song J, Wang J, Zhao L, Huang S, Dissanayake G. Dynamic reconstruction of deformable soft-tissue with stereo scope in minimal invasive surgery. IEEE Robot Autom Lett. 2018;3:155-62.
245. Zhou H, Jagadeesan J. Real-time dense reconstruction of tissue surface from stereo optical video. IEEE Trans Med Imaging. 2020;39:400-12.
246. Zhou H, Jayender J. EMDQ-SLAM: real-time high-resolution reconstruction of soft tissue surface from stereo laparoscopy videos. Med Image Comput Comput Assist Interv. 2021;12904:331-40.
247. Wei G, Feng G, Li H, Chen T, Shi W, Jiang Z. A novel slam method for laparoscopic scene reconstruction with feature patch tracking. In: 2020 International Conference on Virtual Reality and Visualization (ICVRV); 2020 Nov 13-14; Recife, Brazil. IEEE; 2020. pp. 287-91.
248. Wang Y, Long Y, Fan SH, Dou Q. Neural rendering for stereo 3D reconstruction of deformable tissues in robotic surgery. In: Wang L, Dou Q, Fletcher PT, Speidel S, Li S, editors. Medical Image Computing and Computer Assisted Intervention - MICCAI 2022. Cham: Springer; 2022. pp. 431-41.
249. Zha R, Cheng X, Li H, Harandi M, Ge Z. EndoSurf: neural surface reconstruction of deformable tissues with stereo endoscope videos. In: Greenspan H, et al., editors. Medical Image Computing and Computer Assisted Intervention - MICCAI 2023. Cham: Springer; 2023. pp. 13-23.
250. Newcombe RA, Fox D, Seitz SM. Dynamicfusion: reconstruction and tracking of non-rigid scenes in real-time. In: 2015 IEEE Conference on Computer Vision and Pattern Recognition (CVPR); 2015 Jun 7-12; Boston, MA, USA. IEEE; 2015. pp. 343-52.
251. Li Y, Richter F, Lu J, et al. SuPer: a surgical perception framework for endoscopic tissue manipulation with surgical robotics. IEEE Robot Autom Lett. 2020;5:2294-301.
252. Mangulabnan JE, Soberanis-Mukul RD, Teufel T, et al. An endoscopic chisel: intraoperative imaging carves 3D anatomical models. Int J Comput Assist Radiol Surg. 2024;19:1359-66.
253. Nguyen KT, Tozzi F, Rashidian N, Willaert W, Vankerschaver J, De Neve W. Towards abdominal 3-D scene rendering from laparoscopy surgical videos using NeRFs. In: Cao X, Xu X, Rekik I, Cui Z, Ouyang X, editors. Machine Learning in Medical Imaging. Cham: Springer; 2024. pp. 83-93.
254. Hein J, Seibold M, Bogo F, et al. Towards markerless surgical tool and hand pose estimation. Int J Comput Assist Radiol Surg. 2021;16:799-808.
255. Félix I, Raposo C, Antunes M, Rodrigues P, Barreto JP. Towards markerless computer-aided surgery combining deep segmentation and geometric pose estimation: application in total knee arthroplasty. Comput Methods Biomech Biomed Eng Imaging Vis. 2021;9:271-8.
256. Li Z, Shu H, Liang R, et al. TAToo: vision-based joint tracking of anatomy and tool for skull-base surgery. Int J Comput Assist Radiol Surg. 2023;18:1303-10.
257. Murphy-Chutorian E, Trivedi MM. Head pose estimation in computer vision: a survey. IEEE Trans Pattern Anal Mach Intell. 2009;31:607-26.
258. Toshev A, Szegedy C. Deeppose: human pose estimation via deep neural networks. In: 2014 IEEE Conference on Computer Vision and Pattern Recognition; 2014 Jun 23-28; Columbus, OH, USA. IEEE; 2014. pp. 1653-60.
259. Allan M, Chang P, Ourselin S, et al. Image based surgical instrument pose estimation with multi-class labelling and optical flow. In: Navab N, Hornegger J, Wells WM, Frangi A, editors. Medical Image Computing and Computer-Assisted Intervention -- MICCAI 2015. Cham: Springer; 2015. pp. 331-8.
260. Peng S, Liu Y, Huang Q, Zhou X, Bao H. Pvnet: pixel-wise voting network for 6DoF pose estimation. In: 2019 IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR); 2019 Jun 15-20; Long Beach, CA, USA. IEEE; 2019. pp. 4556-65.
261. Do TT, Cai M, Pham T, Reid I. Deep-6dpose: Recovering 6d object pose from a single rgb image. arXiv. [Preprint.] Feb 28, 2018 [accessed 2024 Jul 3]. Available from: https://arxiv.org/abs/1802.10367.
262. He Z, Feng W, Zhao X, Lv Y. 6D pose estimation of objects: recent technologies and challenges. Appl Sci. 2021;11:228.
263. Marullo G, Tanzi L, Piazzolla P, Vezzetti E. 6D object position estimation from 2D images: a literature review. Multimed Tools Appl. 2023;82:24605-43.
264. Hasson Y, Tekin B, Bogo F, Laptev I, Pollefeys M, Schmid C. Leveraging photometric consistency over time for sparsely supervised hand-object reconstruction. In: 2020 IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR); 2020 Jun 13-19; Seattle, WA, USA. IEEE; 2020. pp. 568-77.
265. Kadkhodamohammadi A, Gangi A, de Mathelin M, Padoy N. Articulated clinician detection using 3D pictorial structures on RGB-D data. Med Image Anal. 2017;35:215-24.
266. Padoy N. Machine and deep learning for workflow recognition during surgery. Minim Invasive Ther Allied Technol. 2019;28:82-90.
267. Kadkhodamohammadi A, Gangi A, de Mathelin M, Padoy N. A multi-view rgb-d approach for human pose estimation in operating rooms. In: 2017 IEEE Winter Conference on Applications of Computer Vision (WACV); 2017 Mar 24-31; Santa Rosa, CA, USA. IEEE; 2017. pp. 363-72.
268. Long Y, Wei W, Huang T, Wang Y, Dou Q. Human-in-the-loop embodied intelligence with interactive simulation environment for surgical robot learning. IEEE Robot Autom Lett. 2023;8:4441-8.
269. Killeen BD, Cho SM, Armand M, Taylor RH, Unberath M. In silico simulation: a key enabling technology for next-generation intelligent surgical systems. Prog Biomed Eng. 2023;5:032001.
270. Munawar A, Wang Y, Gondokaryono R, Fischer GS. A real-time dynamic simulator and an associated front-end representation format for simulating complex robots and environments. In: 2019 IEEE/RSJ International Conference on Intelligent Robots and Systems (IROS); 2019 Nov 3-8; Macau, China. IEEE; 2019. pp. 1875-82.
271. Munawar A, Li Z, Kunjam P, et al. Virtual reality for synergistic surgical training and data generation. Comput Methods Biomech Biomed Eng Imaging Vis. 2022;10:366-74.
272. Munawar A, Li Z, Nagururu N, et al. Fully immersive virtual reality for skull-base surgery: surgical training and beyond. Int J Comput Assist Radiol Surg. 2024;19:51-9.
273. Ishida H, Barragan JA, Munawar A, et al. Improving surgical situational awareness with signed distance field: a pilot study in virtual reality. In: 2023 IEEE/RSJ International Conference on Intelligent Robots and Systems (IROS); 2023 Oct 1-5; Detroit, MI, USA. IEEE; 2023. pp. 8474-9.
274. Ishida H, Sahu M, Munawar A, et al. Haptic-assisted collaborative robot framework for improved situational awareness in skull base surgery. arXiv. [Preprint.] Jan 22, 2024 [accessed 2024 Jul 3]. Available from: https://arxiv.org/abs/2401.11709.
275. Sahu M, Ishida H, Connolly L, et al. Integrating 3D slicer with a dynamic simulator for situational aware robotic interventions. arXiv. [Preprint.] Jan 22, 2024 [accessed 2024 Jul 3]. Available from: https://arxiv.org/abs/2401.11715.
276. Su YH, Munawar A, Deguet A, et al. Collaborative robotics toolkit (crtk): open software framework for surgical robotics research. In: 2020 Fourth IEEE International Conference on Robotic Computing (IRC); 2020 Nov 9-11; Taichung, Taiwan. IEEE; 2020. pp. 48-55.
277. Fedorov A, Beichel R, Kalpathy-Cramer J, et al. 3D slicer as an image computing platform for the quantitative imaging network. Magn Reson Imaging. 2012;30:1323-41.
278. Shi Y, Deng X, Tong Y, et al. Synergistic digital twin and holographic augmented-reality-guided percutaneous puncture of respiratory liver tumor. IEEE Trans Human-Mach Syst. 2022;52:1364-74.
279. Poletti G, Antonini L, Mandelli L, et al. Towards a digital twin of coronary stenting: a suitable and validated image-based approach for mimicking patient-specific coronary arteries. Electronics. 2022;11:502.
280. Aubert K, Germaneau A, Rochette M, et al. Development of digital twins to optimize trauma surgery and postoperative management. A case study focusing on tibial plateau fracture. Front Bioeng Biotechnol. 2021;9:722275.
281. Hernigou P, Olejnik R, Safar A, Martinov S, Hernigou J, Ferre B. Digital twins, artificial intelligence, and machine learning technology to identify a real personalized motion axis of the tibiotalar joint for robotics in total ankle arthroplasty. Int Orthop. 2021;45:2209-17.
282. Shinozuka K, Turuda S, Fujinaga A, et al. Artificial intelligence software available for medical devices: surgical phase recognition in laparoscopic cholecystectomy. Surg Endosc. 2022;36:7444-52.
283. Funke I, Mees ST, Weitz J, Speidel S. Video-based surgical skill assessment using 3D convolutional neural networks. Int J Comput Assist Radiol Surg. 2019;14:1217-25.
284. Hashimoto DA, Rosman G, Witkowski ER, et al. Computer vision analysis of intraoperative video: automated recognition of operative steps in laparoscopic sleeve gastrectomy. Ann Surg. 2019;270:414-21.
285. Killeen BD, Zhang H, Wang LJ, et al. Stand in surgeon’s shoes: virtual reality cross-training to enhance teamwork in surgery. Int J Comput Assist Radiol Surg. 2024;19:1213-22.
286. Vercauteren T, Unberath M, Padoy N, Navab N. CAI4CAI: the rise of contextual artificial intelligence in computer assisted interventions. Proc IEEE Inst Electr Electron Eng. 2020;108:198-214.
287. Li Z, Drenkow N, Ding H, et al. On the sins of image synthesis loss for self-supervised depth estimation. arXiv. [Preprint.] Oct 10, 2021 [accessed 2024 Jul 3]. Available from: https://arxiv.org/abs/2109.06163.
288. Gonzales A, Guruswamy G, Smith SR. Synthetic data in health care: a narrative review. PLOS Digit Health. 2023;2:e0000082.
Cite This Article
How to Cite
Download Citation
Export Citation File:
Type of Import
Tips on Downloading Citation
Citation Manager File Format
Type of Import
Direct Import: When the Direct Import option is selected (the default state), a dialogue box will give you the option to Save or Open the downloaded citation data. Choosing Open will either launch your citation manager or give you a choice of applications with which to use the metadata. The Save option saves the file locally for later use.
Indirect Import: When the Indirect Import option is selected, the metadata is displayed and may be copied and pasted as needed.







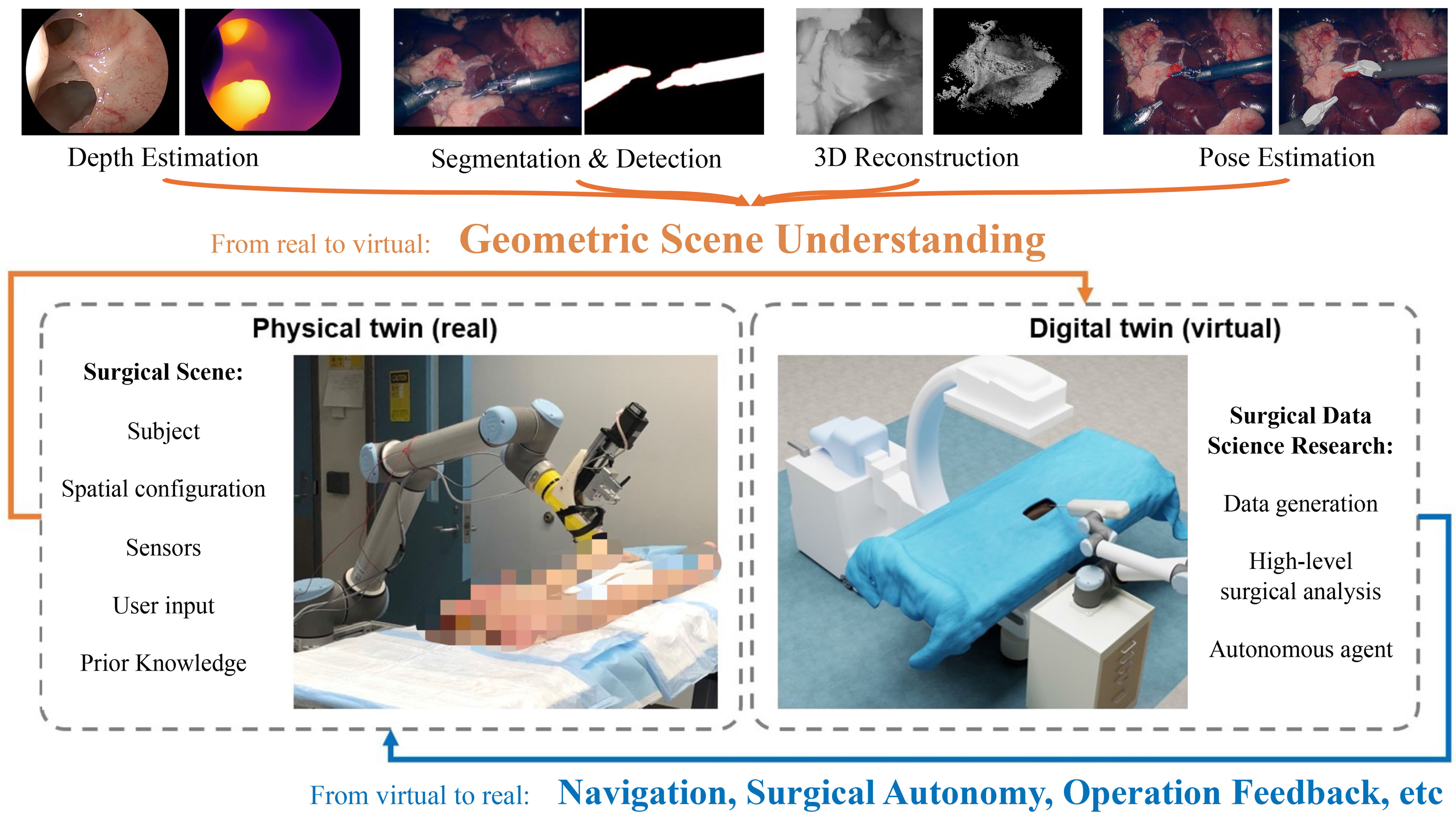
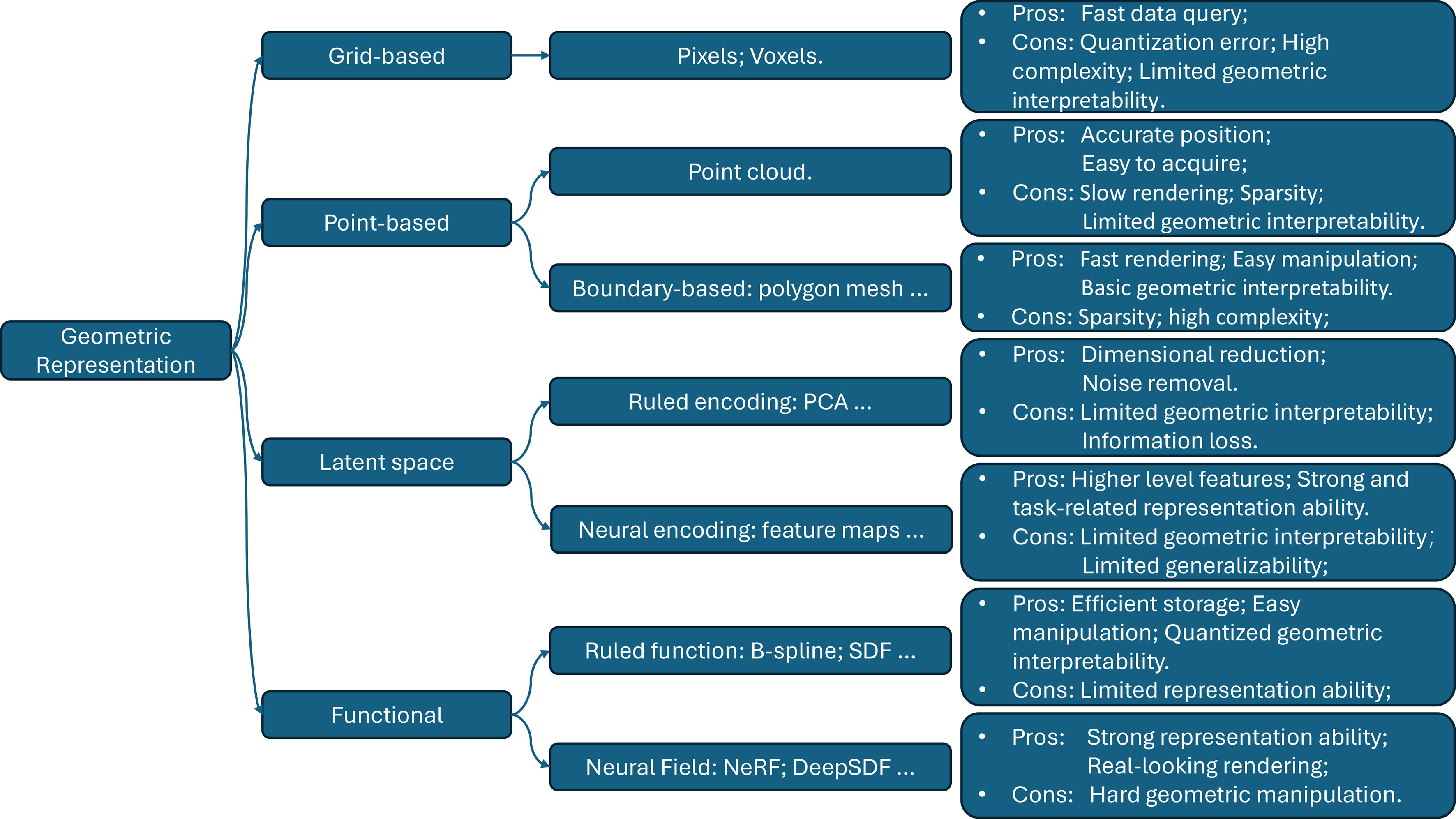
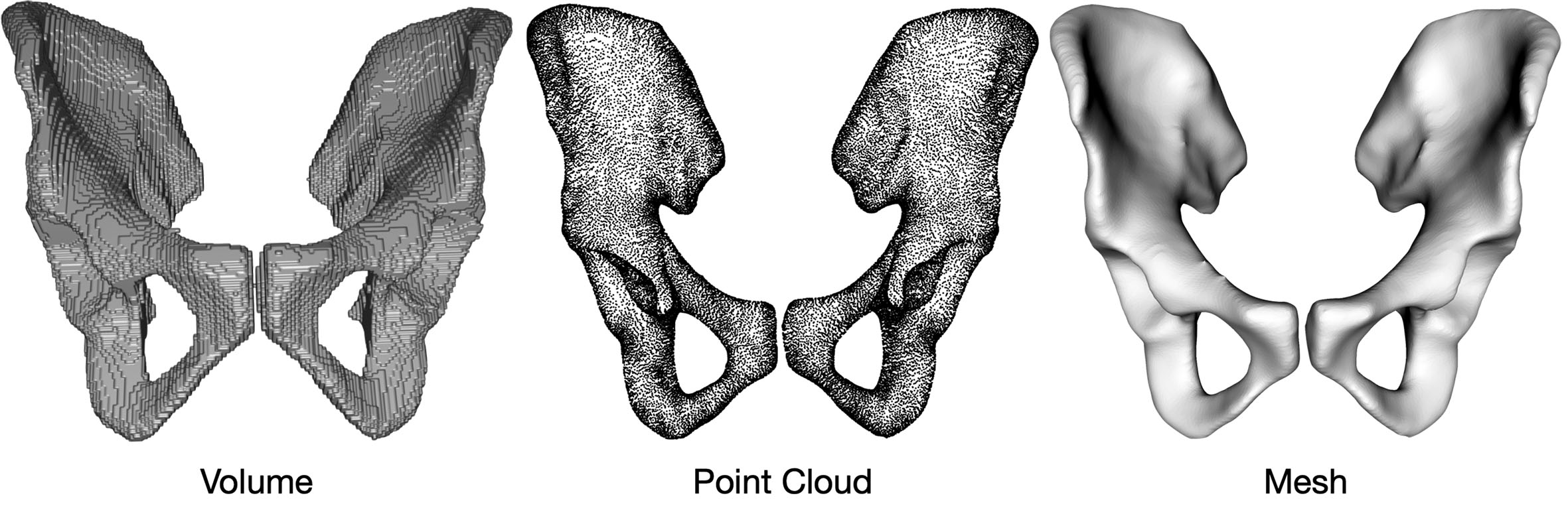
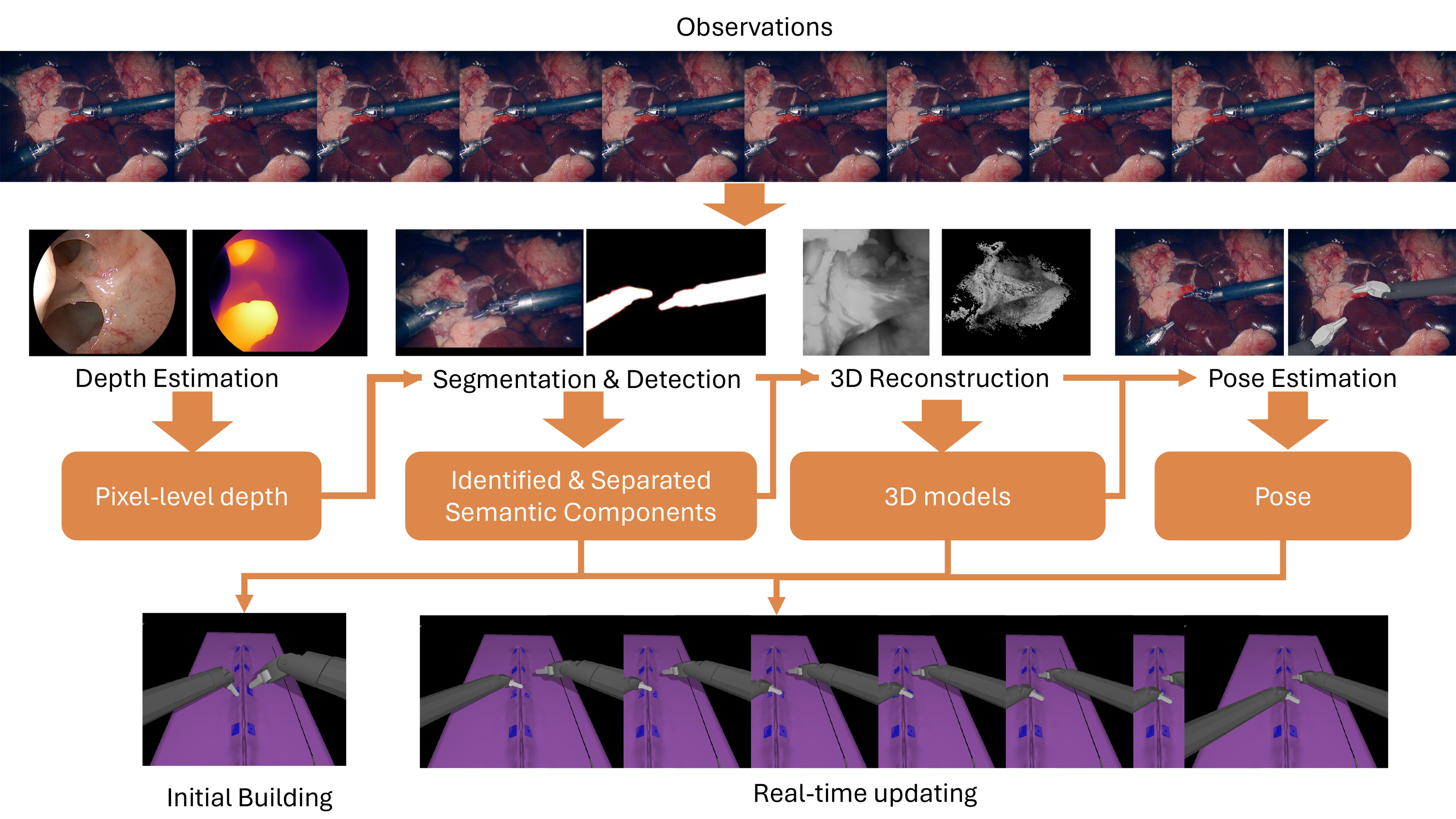













Comments
Comments must be written in English. Spam, offensive content, impersonation, and private information will not be permitted. If any comment is reported and identified as inappropriate content by OAE staff, the comment will be removed without notice. If you have any queries or need any help, please contact us at [email protected].