Lactobacillus, glycans and drivers of health in the vaginal microbiome
Abstract
A microbiome consists of microbes and their genomes, encompassing bacteria, viruses, fungi, protozoa, archaea, and eukaryotes. These elements interact dynamically in the specific environment in which they reside and evolve. In the past decade, studies of various microbiomes have been prevalent in the scientific literature, accounting for the shift from culture-dependent to culture-independent identification of microbes using new high-throughput sequencing technologies that decipher their composition and sometimes provide insights into their functions. Despite tremendous advances in understanding the gut microbiome, relatively little attention has been devoted to the vaginal environment, notably regarding the ubiquity and diversity of glycans which denote the significant role they play in the maintenance of homeostasis. Hopefully, emerging technologies will aid in the determination of what is a healthy vaginal microbiome, and provide insights into the roles of Lactobacillus, glycans and microbiome-related drivers of health and disease.
Keywords
THE VAGINAL MICROBIOME
The human vaginal microbiome comprises a diverse set of organisms that can associate with health or disease and vary across populations[1-3]. It is a complex ecosystem, varying over the course of a woman’s life, constantly fluctuating during the menstrual cycle[4]. Historically, five community state types (CSTs) have been used to describe the vaginal microbiome, encompassing four Lactobacillus-dominant communities that primarily consist of L. crispatus, L. gasseri, L. jensenii, and L. iners, and one non-Lactobacillus dominant diverse community[5]. Altogether, the Lactobacillus-dominated groups occur in approximately 70% of women[6]. The non-Lactobacillus dominant CST typically comprises Gardnerella, Prevotella, Sneathia, Atopobium, Molibuncus, Clostridium, Corynebacterium, Staphylococcus, Streptococcus, Enterococcus, and Mycoplasma[3,7-10]. Although a diversity of gut microbes is typically associated with health, it is not the case in vaginal microbiomes. Indeed, most vaginal microbial populations are dominated by a single genus, Lactobacillus, characterized by Gram-positive anaerobic or microaerophilic rods with peptidoglycan cell walls[11]. This genus is commonly associated with better clinical outcomes[12]. Although diverse Lactobacillus species are associated with a healthy vaginal microbiome, they all usually produce D- and L-lactic acid, which is inhibitory to pathogens and creates anti-inflammatory conditions[13-15]. However, some species like L. iners produce moderate amounts of L-lactic acid, but do not produce the D-isomer[14-16]. Besides their biochemical attributes, structural elements on the bacterial surface also contribute to the microbe-host molecular dialogue[17]. Indeed, microbial recognition hinges on the presentation of cell surface components, especially glycans that interact with host epithelial and immune cells[17-20]. The surface composition differs across species such as L. gasseri, L. jensenii, L. iners and L. crispatus, and others, notably with regard to S-layers. Of note, L. crispatus is the only characterized vaginal Lactobacillus that produces an S-layer, which is comprised of non-covalently bound “crystalline arrays of self-assembling proteins found outermost on the cell wall”[11].
SURFACE COMPONENTS
The S-layer and S-layer-associated proteins (SLAPs) of Lactobacillus acidophilus have been studied and characterized most extensively amongst lactobacilli[11,21]. Such S-layers are associated with cell shape, enhanced adherence to host epithelial cells, and immunomodulatory responses[11,22]. Dendritic cells (DCs) are involved in molecular pattern recognition through sensors like toll-like receptors (TLRs). Studies involving S-layer proteins of the widely commercialized probiotic strain L. acidophilus NCFM[23] have implicated SlpA, SlpB, and SlpX in immunomodulation. In particular, SlpA has been shown to interact with receptors on antigen-presenting DCs, which are important sentinels for mucosal surfaces[24]. Likewise, the interaction between NCFM and DC-SIGN (Dendritic Cell-Specific ICAM-3 intercellular adhesion molecule grabbing non-integrin)[17] receptor drives the production of anti-inflammatory IL-10, whereas the interaction of an NCFM mutant with a chromosomal inversion over-expressing slpB and under-expressing slpA led to the production of proinflammatory cytokines[24]. The homeostatic anti-inflammatory properties of SlpA were shown to mitigate murine colitis[25] and also act via DCs to trigger signaling pathways that inhibit viral infections[26].
The S-layer of L. acidophilus provides a scaffold for numerous SLAPs that are secreted, non-covalently bound[21], and display important surface features[11,27]. Hymes et al.[28] characterized a SLAP binding to fibronectin, and Johnson and Klaenhammer[29] described another SLAP, the AcmB autolysin, which is involved with in vitro binding to mucin and the extracellular matrix proteins fibronectin, collagen, and laminin. The extracellular matrix is a network of molecules produced by resident cells, providing structural support for cells and tissues[30] and regulating cell signaling and adhesion[31]. There are also reports exploring the S-layer and S-layer associated proteins of Lactobacillus crispatus. Antikainen et al.[32-34] demonstrated that S-layer proteins of L. crispatus adhere to collagen, and laminin in the context of intestinal cells. An S-layer producing vaginal L. crispatus isolate was highly adherent to cervicovaginal epithelial cells and was antagonistic to pathogens of the genitourinary tract[35,36]. In silico analyses showed the presence of AcmB orthologs in other S-layer-producing Lactobacillus[29]. When comparing L. crispatus genomes, Pan et al.[37] found heterogeneity regarding the presence of autolysin and acmB-type genes in isolates, though no clear association with a particular isolation source was observed. Furthermore, Tytgat and Lebeer[38] highlight that bacterial glycoconjugates at the microbial surface, including S-layers, may be glycosylated. In fact, microbial glycans comprise much of the bacterial cell surface[39]. To date, S-layer glycans have only been confirmed in L. buchneri and L. kefir[40]. Thus, future studies should determine whether L. acidophilus and other Lactobacillus S-layer proteins are glycosylated[41].
GLYCOSYLATION AND THE GLYCOME
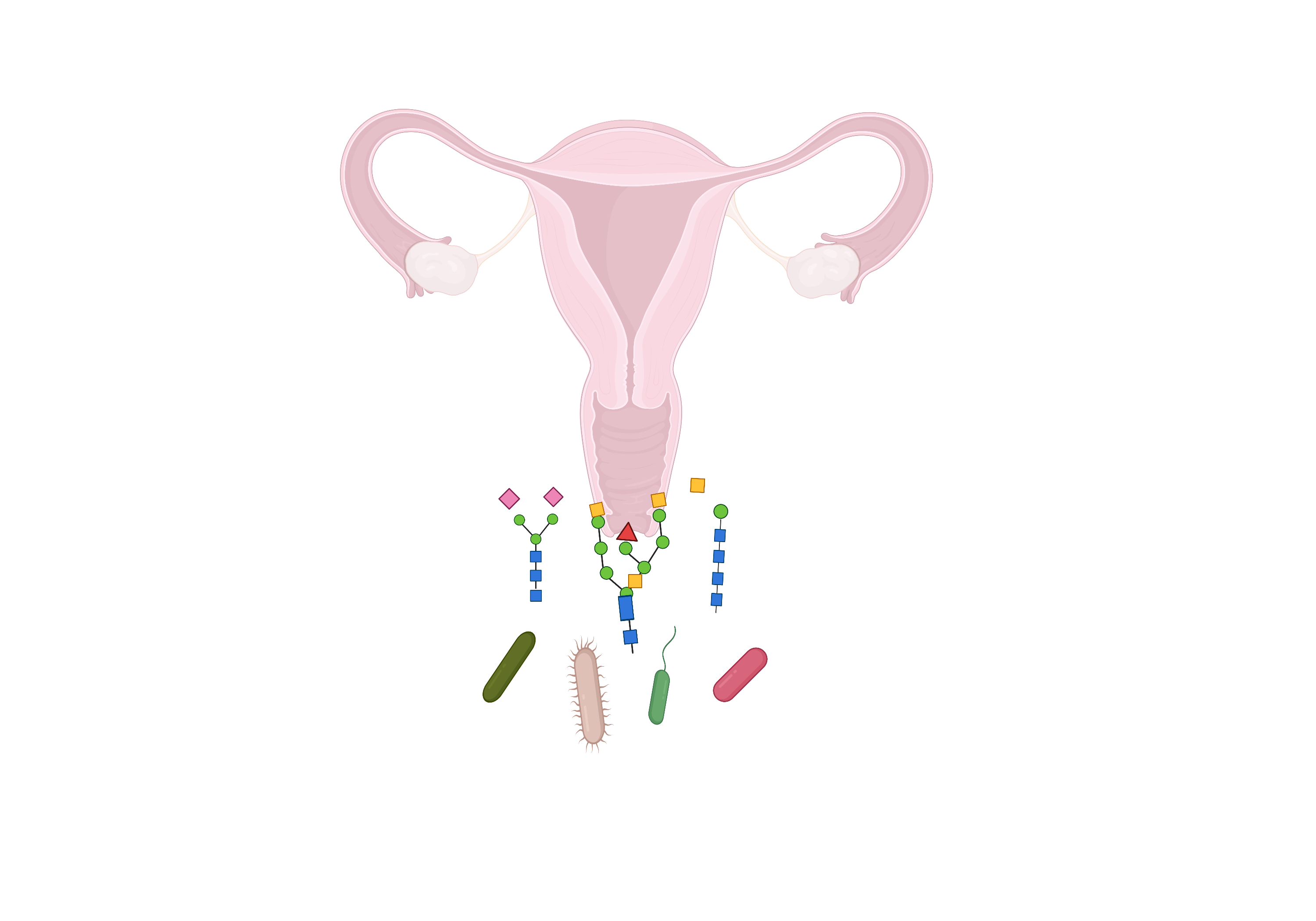
The glycome is the entirety of a cell’s carbohydrates, either free or as moieties of glycoconjugated macromolecules. Almost all cells are surrounded by glycans, forming a “sugar jacket” comprising proteoglycans, glycosphingolipids, and glycoproteins that form a glycocalyx[42]. In vertebrates, mucosal glycan chains typically terminate in various sialic acid molecules[43,44]. Within the glycome, the negatively charged sialome[45] plays a role in signaling by concealing antigens on cell surfaces, which consequently appear as “self”, thereby weakening immunoreactivity[46]. The diverse functions of glycans include structural modularity with various glycoconjugates, and providing specificity for glycan-binding proteins and receptors[30,42,47]. Both receptors and ligands may contain essential glycan domains, such as pattern-recognition receptors, encompassing TLRs which are transmembrane glycoproteins that have evolved to recognize conserved molecular patterns on microbial surfaces, typically referred to as MAMPs (Microorganism-Associated Microbial Patterns) that may also contain glycans[17,18,48]. Glycosylation is an essential regulatory mechanism for post-translational processing, which plays a crucial role in the assignment of protein structure, function, and stability, especially 3-D conformation. This in turn influences protein-protein interactions like signaling[42] as well as eukaryotic viral and bacteriophage attachment[49]. Some Interleukins, cytokines, viral coat proteins, and G protein-coupled receptors are glycosylated, as well as immunoglobulins and hormones such as gonadotropins, luteinizing hormone, follicle-stimulating hormone, and thyroid-stimulating hormone[50-52]. “Essentially all surface-localized immune receptors are glycoproteins”[53]. Glycans are integral for immune system modulation and interaction with cells such as macrophages, monocytes, natural killer cells, antigen-presenting cells like dendritic cells, and T cells. These interactions impact both innate and adaptive immunities, cytokine production, and epithelial cell responses[18,53]. Glycans can exert dual immune roles, acting either in an inhibitory or stimulatory manner[18,19], and can be involved in tolerance or autoimmunity[18-20].
On the host side, the vaginal mucus is also highly glycosylated. It is composed of glycoprotein mucins, other secreted proteins like immunoglobulins[54], and antimicrobial peptides produced by mucosal epithelial cells and neutrophils, some of which bind glycans[55,56]. The branched carbohydrate moieties can make up to 80% of mucin weight[54] and usually terminate in an outermost sialic acid residue[57]. Mucus sialoglycoproteins are believed to entrap microorganisms, protecting epithelial cells against infection. Cell surface mucins can bind pathogens, indicating a role of mucus in the innate immunity of the female genital tract[12,57,58]. Consequently, the glycome status of host cells, commensal microbes, and mucosal surfaces in a microbiome are important in the maintenance of homeostasis and health, implying that glycome disruption could lead to dysbiosis.
BACTERIAL VAGINOSIS
In the context of women’s health, bacterial vaginosis (BV) is a prevalent form of vaginal dysbiosis associated with a variety of adverse health outcomes[59]. It is often found in non-Lactobacillus-dominated microbiomes[3,4,10]. Biofilm presence on vaginal epithelial cells is an important factor in the evolution of BV and explains episodes of recurrent infections[60]. A signature of BV is the presence of a dense polymicrobial biofilm on the vaginal surface believed to be initiated by Gardnerella vaginalis, providing a scaffold for other species to adhere to[10]. An under-reported factor in biofilm formation is glycosyltransferase activity, as well as the destructive action of sialidases on the protective mucosal glycan surfaces of vaginal epithelia, which possibly facilitates adhesion of vaginal pathogens such as Gardnerella and Prevotella[61,62].
Moncla et al.[54] have shown that glycosidase and sialidase activity is associated with a reduced number of sialic acid binding sites, a virulence factor in pathogens of mucosal surfaces, and a feature of BV[58]. Two bacteria typically associated with BV are Gardnerella vaginalis and Prevotella bivia. Sialidase is produced by many P. bivia isolates, but only by 25% of G. vaginalis isolates. The sialidase produced by Prevotella is cell-bound, whereas Gardnerella sialidases are extracellular and affect the vaginal environment differently[54,58,62]. When sialidases are secreted, they may remove sialic acid residues from carbohydrate chains distant from the organism[63], and subsequently affect the function of cells and molecules like immunoglobulins. This would make sialic acids available to other organisms capable of their catabolism and furthermore, expose the “open” carbohydrate chains to exo- and endo-glycosidase attack as well as other hydrolytic enzymes like mucinases, sulfatases, proline dipeptidases, and fucosidases[19,58,64-66].
HOST-MICROBE INTERACTIONS
In their insightful opinion, Tytgat and de Vos[17] equate the “array of glycoconjugates on bacterial surfaces as strain-specific barcodes generating diversity as ligands for shaping microbial-host interactions”[17]. Even though the field of bacterial glycobiology is expanding, the scarcity of studies on bacterial cell surface protein glycosylation in general, and in the context of women’s health is perplexing. Sun et al.[71] carried out a comparative genomic analysis of 213 lactobacilli, and highlighted the diversity of glycotransferases documented. However, surface glycoconjugates cannot be inferred genetically, since they are post-translational modifications, highlighting the need for functional and biochemical characterization[23]. Likewise, genomic studies of L. gasseri[72] and L. jensenii[73] have not substantiated their beneficial roles in the vaginal microbiome. Petrova et al.[6] report that L. jensenii can reduce adherence and invasiveness of N. gonorrheae, and that L. gasseri can displace the gonorrhea coccus, but these traits can vary across strains. A characteristic of some L .crispatus and L. gasseri strains is the presence of genes encoding mucin binding proteins, but additional functional and mechanistic insights are needed.[12] The role of L. iners in contributing to vaginal health or disease is unclear and sometimes subject to controversy[66]. Indeed, L. iners shares attributes with Gardnerella, a pathogen associated with BV, such as: a small genome indicative of symbiotic/parasitic lifestyle, moderate lactic acid production[6,14,15], secretion of a cholesterol-dependent cytolysin, and overgrowth during menstruation[10,16]. Some believe that L. iners may be a transitional organism between health and dysbiosis[10,74]. Actually, L. iners, is found in low to moderate abundance in the non-Lactobacillus-dominated vaginal microbiome[5,65].
A bacterial surface glycoconjugate barcode sends a molecular message to other community members[17]. A critical step in deciphering these barcoded messages is to determine the glycosylation of surface proteins in vaginal lactobacilli of interest. This can be achieved by cell shaving with trypsin[75] or by Lithium chloride extraction of S-layer proteins and SLAPs[11,27] and determination of glycosylation using lectin microarrays, monoclonal antibodies, synthetic glycans, or prediction tools[39,74,76,77]. In particular, it would be interesting to determine L. crispatus surface glycosylation given the association of this species with vaginal health and homeostasis[78]. Besides, L. crispatus isolates from vaginal, intestinal, and poultry sources display very different S-layer and SLAP profiles, providing a unique opportunity to determine glycome diversity across environments for one species[21].
Glycogen is a major carbohydrate source due to its cyclical release from vaginal epithelial cells. Many commensals rely on host amylases to break it down into usable sugars. Van der Veer et al.[79] 2019 found some L. crispatus isolates with intact pullulanase type 1 genes, enabling them to directly utilize glycogen, which would provide a competitive advantage. Genomes of L. crispatus encode diverse hypervariable content, including prophages, autolysins, bacteriocins, and various systems related to mobile genetic elements such as plasmid stabilization systems, toxin-antitoxin systems, and CRISPR (Clustered Regularly Interspaced Short Palindromic Repeat) and associated sequences CRISPR-Cas immune systems[37,80]. Studies have reported genomic islands encoding enzymes involved in exopolysaccharide (EPS) production on plasmids in L. crispatus, which could enhance adherence, biofilm formation, and exclusion of pathogens[79]. Yet, it is unclear how the immunological L. crispatus “message” differs between S-layer presentation and EPS, and whether L. crispatus alters its surface glycome via EPS. It would be illuminating to determine the glycome of all major vaginal microbiome bacteria. Since many components of extracellular matrices are glycosylated, the ECM of the vaginal microbiome should be explored[31,81]. Importantly, the effects of other factors in play should also be determined, notably the virome[49] mycobiome[82], hormones[13,58], metabolites[7,83], stress[14], male factor transfers[3], sexual partners[6], douching[58,84], and endocrine disruptors[85]. Unfortunately, since the human cervico-vaginal environment possesses unique attributes such as a particularly low pH, due to Lactobacillus lactic acid production[6,15], there is no adequate animal model. Nevertheless, we could exploit rising technologies such as “organ-on-a-chip”[86] or 3D EpiVaginal tissueTM[87] for future studies.
CONCLUSIONS
Despite tremendous advances in the study of the intestinal microbiome, the relative paucity of studies on the vaginal microbiome is puzzling. A deeper and more comprehensive understanding of microbial dynamics and bacterial functions in the vaginal microbiome would drive the development of novel products to maintain, enhance or restore vaginal health and prevent or treat dysbiosis. This could also enable the development of biomarkers to detect microbiome aberrations and diagnose unhealthy conditions[6]. Historically, our limited understanding has also been hampered by the lack of glycoscience-related tools, though recent efforts are encouraging, promoted by the Consortium of Functional Glycomics and international entities such as EuroCarb and the Japanese Consortium for Glycobiology and Glycotechnology[53].
McKitrick et al.[39] summarized topics covered at a recent NIH workshop entitled “Glycoscience and Immunology at the Crossroads of Biology.” They present a Venn diagram where immunology, microbiology, and glycobiology overlap to encompass glycoscience, infection, and immunity. Participants discussed the need to determine glycan structures, linkages, stereochemical orientation, and functionality[39], as widespread essential factors with variable chain length, linkage, and branching with inherent functional differences[88]. Given the implication of glycans in cell activation, differentiation, and development, a deeper understanding of their role in women’s health would be beneficial[42,89]. Glycome studies will complement genomic and functional analyses of the vaginal microbiome [Figure 1] and reveal the importance of glycans in other microbiomes. This will open new avenues to manipulate the composition and function of key bacterial species driving women’s health and disease.
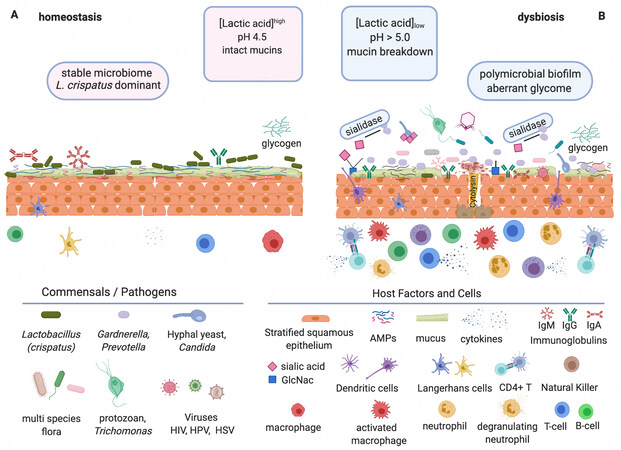
Figure 1. Vaginal homeostasis vs. dysbiosis. Glycobiology is key to understanding interactions within the vaginal microbiome since many elements encompassing the microbiota and the host are glycosylated or bind glycans. The immune state is affected[90] in different ways between a healthy state of homeostasis (A) and a disease state of symbiosis, which in turn contributes to either health (A) or dysbiosis (B) characterized by distinct commensals and pathogens interacting with host factors and cells. (A) Homeostasis. Lactobacillus crispatus is deemed to be the preferred vaginal microbiome commensal when dominant due to its high lactic acid production, from glycogen degradation, resulting in beneficial low pH. (B) Dysbiosis. This condition does not have the beneficial protective effects of low pH. Presented by multi-species, non-Lactobacillus flora, including pathogens such as Prevotella and Gardnerella. Virulence factors are produced such as: biofilms, hydrolytic enzymes (e.g., sialidases), and cytolysins which can lead to the breakdown of mucins and epithelial cells, disruption of the homeostatic glycome, and immune response (e.g., deglycosylation of immunoglobulins and activation of immune factors). These conditions in turn promote the rise of undesirable members of the microbiome, such as viruses, yeast, and even protozoa[6,91]. Figure created using BioRender.com.
DECLARATIONS
Authors’ contributionsWrote the manuscript: Sanozky-Dawes R
Edited the manuscript: Barrangou R
Availability of data and materialsNot applicable.
Financial support and sponsorshipThe authors acknowledge internal support from North Carolina State University.
Conflicts of interestBoth authors declared that there are no conflicts of interest.
Ethical approval and consent to participateNot applicable.
Consent for publicationNot applicable.
Copyright© The Author(s) 2022.
REFERENCES
1. Stout MJ, Wylie TN, Gula H, Miller A, Wylie KM. The microbiome of the human female reproductive tract. Current Opinion in Physiology 2020;13:87-93.
2. Kho ZY, Lal SK. The human gut microbiome - a potential controller of wellness and disease. Front Microbiol 2018;9:1835.
3. Koedooder R, Mackens S, Budding A, et al. Identification and evaluation of the microbiome in the female and male reproductive tracts. Hum Reprod Update 2019;25:298-325.
4. Chen X, Lu Y, Chen T, Li R. The female vaginal microbiome in health and bacterial vaginosis. Front Cell Infect Microbiol 2021;11:631972.
5. France M, Alizadeh M, Brown S, Ma B, Ravel J. Towards a deeper understanding of the vaginal microbiota. Nat Microbiol 2022;7:367-78.
6. Petrova MI, Lievens E, Malik S, Imholz N, Lebeer S. Lactobacillus species as biomarkers and agents that can promote various aspects of vaginal health. Front Physiol 2015;6:81.
7. Delgado-Diaz DJ, Tyssen D, Hayward JA, Gugasyan R, Hearps AC, Tachedjian G. Distinct immune responses elicited from cervicovaginal epithelial cells by lactic acid and short chain fatty acids associated with optimal and non-optimal vaginal microbiota. Front Cell Infect Microbiol 2019;9:446.
8. Kaambo E, Africa C, Chambuso R, Passmore JS. Vaginal microbiomes associated with aerobic vaginitis and bacterial vaginosis. Front Public Health 2018;6:78.
10. Petrova MI, Reid G, Vaneechoutte M, Lebeer S. Lactobacillus iners: friend or foe? Trends Microbiol 2017;25:182-91.
11. Johnson B, Selle K, O’Flaherty S, Goh YJ, Klaenhammer T. Identification of extracellular surface-layer associated proteins in Lactobacillus acidophilus NCFM. Microbiology (Reading) 2013;159:2269-82.
12. Vagios S, Mitchell CM. Mutual preservation: a review of interactions between cervicovaginal mucus and microbiota. Front Cell Infect Microbiol 2021;11:676114.
13. Gliniewicz K, Schneider GM, Ridenhour BJ, et al. Comparison of the vaginal microbiomes of premenopausal and postmenopausal women. Front Microbiol 2019;10:193.
14. Amabebe E, Anumba DOC. The vaginal microenvironment: the physiologic role of Lactobacilli. Front Med (Lausanne) 2018;5:181.
15. Witkin SS, Linhares IM. Why do Lactobacilli dominate the human vaginal microbiota? BJOG 2017;124:606-11.
17. Tytgat HLP, de Vos WM. Sugar coating the envelope: glycoconjugates for microbe-host crosstalk. Trends Microbiol 2016;24:853-61.
18. Pereira MS, Alves I, Vicente M, et al. Glycans as key checkpoints of T cell activity and function. Front Immunol 2018;9:2754.
19. Clark GF, Schust DJ. Manifestations of immune tolerance in the human female reproductive tract. Front Immunol 2013;4:26.
20. Rabinovich GA, Toscano MA. Turning ‘sweet’ on immunity: galectin-glycan interactions in immune tolerance and inflammation. Nat Rev Immunol 2009;9:338-52.
21. Johnson BR, Hymes J, Sanozky-Dawes R, Henriksen ED, Barrangou R, Klaenhammer TR. Conserved S-layer-associated proteins revealed by exoproteomic survey of S-layer-forming Lactobacilli. Appl Environ Microbiol 2016;82:134-45.
22. Hymes JP, Klaenhammer TR. Stuck in the middle: fibronectin-binding proteins in gram-positive bacteria. Front Microbiol 2016;7:1504.
23. Lin B, Qing X, Liao J, Zhuo K. Role of protein glycosylation in host-pathogen interaction. Cells 2020;9:1022.
24. Konstantinov SR, Smidt H, de Vos WM, et al. S layer protein A of Lactobacillus acidophilus NCFM regulates immature dendritic cell and T cell functions. Proc Natl Acad Sci U S A 2008;105:19474-9.
25. Lightfoot YL, Selle K, Yang T, et al. SIGNR3-dependent immune regulation by Lactobacillus acidophilus surface layer protein A in colitis. EMBO J 2015;34:881-95.
26. Acosta M, Geoghegan EM, Lepenies B, Ruzal S, Kielian M, Martinez MG. Surface (S) layer proteins of Lactobacillus acidophilus block virus infection via DC-SIGN interaction. Front Microbiol 2019;10:810.
27. Klotz C, Goh YJ, O'Flaherty S, Barrangou R. S-layer associated proteins contribute to the adhesive and immunomodulatory properties of Lactobacillus acidophilus NCFM. BMC Microbiol 2020;20:248.
28. Hymes JP, Johnson BR, Barrangou R, Klaenhammer TR. Functional analysis of an S-layer-associated fibronectin-binding protein in Lactobacillus acidophilus NCFM. Appl Environ Microbiol 2016;82:2676-85.
29. Johnson BR, Klaenhammer TR. AcmB is an S-layer-associated β-N-acetylglucosaminidase and functional autolysin in Lactobacillus acidophilus NCFM. Appl Environ Microbiol 2016;82:5687-97.
31. Karamanos NK, Theocharis AD, Piperigkou Z, et al. A guide to the composition and functions of the extracellular matrix. FEBS J 2021;288:6850-912.
32. Antikainen J, Anton L, Sillanpää J, Korhonen TK. Domains in the S-layer protein CbsA of Lactobacillus crispatus involved in adherence to collagens, laminin and lipoteichoic acids and in self-assembly. Mol Microbiol 2002;46:381-94.
33. Sillanpää J, Martínez B, Antikainen J, et al. Characterization of the collagen-binding S-layer protein CbsA of Lactobacillus crispatus. J Bacteriol 2000;182:6440-50.
34. Sun Z, Kong J, Hu S, Kong W, Lu W, Liu W. Characterization of a S-layer protein from Lactobacillus crispatus K313 and the domains responsible for binding to cell wall and adherence to collagen. Appl Microbiol Biotechnol 2013;97:1941-52.
35. Abramov V, Khlebnikov V, Kosarev I, et al. Probiotic properties of Lactobacillus crispatus 2029: homeostatic interaction with cervicovaginal epithelial cells and antagonistic activity to genitourinary pathogens. Probiotics Antimicrob Proteins 2014;6:165-76.
36. Abramov VM, Kosarev IV, Priputnevich TV, et al. S-layer protein 2 of Lactobacillus crispatus 2029, its structural and immunomodulatory characteristics and roles in protective potential of the whole bacteria against foodborne pathogens. Int J Biol Macromol 2020;150:400-12.
37. Pan M, Hidalgo-Cantabrana C, Barrangou R. Host and body site-specific adaptation of Lactobacillus crispatus genomes. NAR Genom Bioinform 2020;2:lqaa001.
38. Tytgat HL, Lebeer S. The sweet tooth of bacteria: common themes in bacterial glycoconjugates. Microbiol Mol Biol Rev 2014;78:372-417.
39. McKitrick TR, Ackerman ME, Anthony RM, et al. The crossroads of glycoscience, infection, and immunology. Front Microbiol 2021;12:731008.
40. Hynönen U, Palva A. Lactobacillus surface layer proteins: structure, function and applications. Appl Microbiol Biotechnol 2013;97:5225-43.
41. Fina Martin J, Palomino MM, Cutine AM, et al. Exploring lectin-like activity of the S-layer protein of Lactobacillus acidophilus ATCC 4356. Appl Microbiol Biotechnol 2019;103:4839-57.
42. Varki A, Gagneux P. Biological functions of glycans. In: Varki A, Cummings RD, Esko JD, et al., editors. Essentials of glycobiology [Internet]. 3rd ed. Cold Spring Harbor (NY): Cold Spring Harbor Laboratory Press; 2015-2017. Chapter 7.
43. Schauer R. Sialic acids as regulators of molecular and cellular interactions. Curr Opin Struct Biol 2009;19:507-14.
46. Lübbers J, Rodríguez E, van Kooyk Y. Modulation of immune tolerance via siglec-sialic acid interactions. Front Immunol 2018;9:2807.
47. Cummings RD, Schnaar RL, Esko JD, Drickamer K, Taylor ME. Principles of glycan recognition. In: Varki A, Cummings RD, Esko JD, et al., editors. Essentials of glycobiology [Internet]. 3rd ed. Cold Spring Harbor (NY): Cold Spring Harbor Laboratory Press; 2015-2017. Chapter 29.
48. Mariano VS, Zorzetto-Fernandes AL, da Silva TA, et al. Recognition of TLR2 N-glycans: critical role in ArtinM immunomodulatory activity. PLoS One 2014;9:e98512.
49. Simpson DJ, Sacher JC, Szymanski CM. Exploring the interactions between bacteriophage-encoded glycan binding proteins and carbohydrates. Curr Opin Struct Biol 2015;34:69-77.
50. Bousfield GR, May JV, Davis JS, Dias JA, Kumar TR. In vivo and in vitro impact of carbohydrate variation on human follicle-stimulating hormone function. Front Endocrinol (Lausanne) 2018;9:216.
51. Campo S, Andreone L, Ambao V, Urrutia M, Calandra RS, Rulli SB. Hormonal regulation of follicle-stimulating hormone glycosylation in males. Front Endocrinol (Lausanne) 2019;10:17.
52. Willey KP. An elusive role for glycosylation in the structure and function of reproductive hormones. Hum Reprod Update 1999;5:330-55.
53. Rabinovich GA, van Kooyk Y, Cobb BA. Glycobiology of immune responses. Ann N Y Acad Sci 2012;1253:1-15.
54. Moncla BJ, Chappell CA, Mahal LK, Debo BM, Meyn LA, Hillier SL. Impact of bacterial vaginosis, as assessed by nugent criteria and hormonal status on glycosidases and lectin binding in cervicovaginal lavage samples. PLoS One 2015;10:e0127091.
55. Mahlapuu M, Håkansson J, Ringstad L, Björn C. Antimicrobial peptides: an emerging category of therapeutic agents. Front Cell Infect Microbiol 2016;6:194.
56. Yarbrough VL, Winkle S, Herbst-Kralovetz MM. Antimicrobial peptides in the female reproductive tract: a critical component of the mucosal immune barrier with physiological and clinical implications. Hum Reprod Update 2015;21:353-77.
57. Lewis AL, Lewis WG. Host sialoglycans and bacterial sialidases: a mucosal perspective. Cell Microbiol 2012;14:1174-82.
58. Moncla BJ, Chappell CA, Debo BM, Meyn LA. The effects of hormones and vaginal microflora on the glycome of the female genital tract: cervical-vaginal fluid. PLoS One 2016;11:e0158687.
59. Morrill S, Gilbert NM, Lewis AL. Gardnerella vaginalis as a cause of bacterial vaginosis: appraisal of the evidence from in vivo models. Front Cell Infect Microbiol 2020;10:168.
60. Hardy L, Cerca N, Jespers V, Vaneechoutte M, Crucitti T. Bacterial biofilms in the vagina. Res Microbiol 2017;168:865-74.
61. Castro J, Machado D, Cerca N. Unveiling the role of Gardnerella vaginalis in polymicrobial bacterial vaginosis biofilms: the impact of other vaginal pathogens living as neighbors. ISME J 2019;13:1306-17.
62. Lewis WG, Robinson LS, Gilbert NM, Perry JC, Lewis AL. Degradation, foraging, and depletion of mucus sialoglycans by the vagina-adapted Actinobacterium Gardnerella vaginalis. J Biol Chem 2013;288:12067-79.
63. Smith SB, Ravel J. The vaginal microbiota, host defence and reproductive physiology. J Physiol 2017;595:451-63.
64. Hoang T, Toler E, DeLong K, et al. The cervicovaginal mucus barrier to HIV-1 is diminished in bacterial vaginosis. PLoS Pathog 2020;16:e1008236.
65. France MT, Fu L, Rutt L, et al. Insight into the ecology of vaginal bacteria through integrative analyses of metagenomic and metatranscriptomic data. Genome Biol 2022;23:66.
66. France MT, Rutt L, Narina S, et al. Complete genome sequences of six Lactobacillus iners strains isolated from the human vagina. Microbiol Resour Announc 2020;9:e00234-20.
67. Varki A, Schnaar RL, Schauer R. Sialic acids and other nonulosonic acids. In: Varki A, Cummings RD, Esko JD, et al., editors. Essentials of glycobiology [Internet]. 3rd ed. Cold Spring Harbor (NY): Cold Spring Harbor Laboratory Press; 2015-2017. Chapter 15.
68. Varki NM, Varki A. Diversity in cell surface sialic acid presentations: implications for biology and disease. Lab Invest 2007;87:851-7.
69. Raposo CD, Canelas AB, Barros MT. Human lectins, their carbohydrate affinities and where to find them. Biomolecules 2021;11:188.
70. Koppolu S, Wang L, Mathur A, et al. Vaginal product formulation alters the innate antiviral activity and glycome of cervicovaginal fluids with implications for viral susceptibility. ACS Infect Dis 2018;4:1613-22.
71. Sun Z, Harris HM, McCann A, et al. Expanding the biotechnology potential of lactobacilli through comparative genomics of 213 strains and associated genera. Nat Commun 2015;6:8322.
72. Zhou X, Yang B, Stanton C, et al. Comparative analysis of Lactobacillus gasseri from Chinese subjects reveals a new species-level taxa. BMC Genomics 2020;21:119.
73. Lee S, You HJ, Kwon B, Ko G. Complete genome sequence of Lactobacillus jensenii strain SNUV360, a probiotic for treatment of bacterial vaginosis isolated from the vagina of a healthy Korean woman. Genome Announc 2017;5:e01757-16.
74. Bonnardel F, Haslam SM, Dell A, et al. Proteome-wide prediction of bacterial carbohydrate-binding proteins as a tool for understanding commensal and pathogen colonisation of the vaginal microbiome. NPJ Biofilms Microbiomes 2021;7:49.
75. Espino E, Koskenniemi K, Mato-Rodriguez L, et al. Uncovering surface-exposed antigens of Lactobacillus rhamnosus by cell shaving proteomics and two-dimensional immunoblotting. J Proteome Res 2015;14:1010-24.
76. Campanero-Rhodes MA, Palma AS, Menéndez M, Solís D. Microarray strategies for exploring bacterial surface glycans and their interactions with glycan-binding proteins. Front Microbiol 2019;10:2909.
78. Oliveira de Almeida M, Carvalho R, Figueira Aburjaile F, et al. Characterization of the first vaginal Lactobacillus crispatus genomes isolated in Brazil. PeerJ 2021;9:e11079.
79. van der Veer C, Hertzberger RY, Bruisten SM, et al. Comparative genomics of human Lactobacillus crispatus isolates reveals genes for glycosylation and glycogen degradation: implications for in vivo dominance of the vaginal microbiota. Microbiome 2019;7:49.
80. Mendes-Soares H, Suzuki H, Hickey RJ, Forney LJ. Comparative functional genomics of Lactobacillus spp. reveals possible mechanisms for specialization of vaginal lactobacilli to their environment. J Bacteriol 2014;196:1458-70.
81. Tyagi T, Alarab M, Leong Y, Lye S, Shynlova O. Local oestrogen therapy modulates extracellular matrix and immune response in the vaginal tissue of post-menopausal women with severe pelvic organ prolapse. J Cell Mol Med 2019;23:2907-19.
82. Bradford LL, Ravel J. The vaginal mycobiome: a contemporary perspective on fungi in women’s health and diseases. Virulence 2017;8:342-51.
83. Puebla-Barragan S, Watson E, van der Veer C, et al. Interstrain variability of human vaginal Lactobacillus crispatus for metabolism of biogenic amines and antimicrobial activity against urogenital pathogens. Molecules 2021;26:4538.
84. Gabriel IM, Vitonis AF, Welch WR, Titus L, Cramer DW. Douching, talc use, and risk for ovarian cancer and conditions related to genital tract inflammation. Cancer Epidemiol Biomarkers Prev 2019;28:1835-44.
85. Dunbar B, Patel M, Fahey J, Wira C. Endocrine control of mucosal immunity in the female reproductive tract: impact of environmental disruptors. Mol Cell Endocrinol 2012;354:85-93.
86. Mancini V, Pensabene V. Organs-on-chip models of the female reproductive system. Bioengineering (Basel) 2019;6:103.
87. Hearps AC, Tyssen D, Srbinovski D, et al. Vaginal lactic acid elicits an anti-inflammatory response from human cervicovaginal epithelial cells and inhibits production of pro-inflammatory mediators associated with HIV acquisition. Mucosal Immunol 2017;10:1480-90.
88. Chaichian S, Moazzami B, Sadoughi F, Haddad Kashani H, Zaroudi M, Asemi Z. Functional activities of beta-glucans in the prevention or treatment of cervical cancer. J Ovarian Res 2020;13:24.
89. Ferreira IG, Pucci M, Venturi G, Malagolini N, Chiricolo M, Dall’Olio F. Glycosylation as a main regulator of growth and death factor receptors signaling. Int J Mol Sci 2018;19:580.
90. Zhou JZ, Way SS, Chen K. Immunology of uterine and vaginal mucosae: (trends in immunology 39, 302-314, 2018). Trends Immunol 2018;39:355.
Cite This Article
Export citation file: BibTeX | RIS
OAE Style
Sanozky-Dawes R, Barrangou R. Lactobacillus, glycans and drivers of health in the vaginal microbiome. Microbiome Res Rep 2022;1:18. http://dx.doi.org/10.20517/mrr.2022.03
AMA Style
Sanozky-Dawes R, Barrangou R. Lactobacillus, glycans and drivers of health in the vaginal microbiome. Microbiome Research Reports. 2022; 1(3): 18. http://dx.doi.org/10.20517/mrr.2022.03
Chicago/Turabian Style
Sanozky-Dawes, Rosemary, Rodolphe Barrangou. 2022. "Lactobacillus, glycans and drivers of health in the vaginal microbiome" Microbiome Research Reports. 1, no.3: 18. http://dx.doi.org/10.20517/mrr.2022.03
ACS Style
Sanozky-Dawes, R.; Barrangou R. Lactobacillus, glycans and drivers of health in the vaginal microbiome. Microbiome. Res. Rep. 2022, 1, 18. http://dx.doi.org/10.20517/mrr.2022.03
About This Article
Copyright
Data & Comments
Data
 Cite This Article 11 clicks
Cite This Article 11 clicks











Comments
Comments must be written in English. Spam, offensive content, impersonation, and private information will not be permitted. If any comment is reported and identified as inappropriate content by OAE staff, the comment will be removed without notice. If you have any queries or need any help, please contact us at support@oaepublish.com.